Shwachman-Bodian-Diamond Syndrome Protein
From Proteopedia
(Difference between revisions)
(→'''SBDS Association with RNA Processing''') |
|||
| (12 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | < | + | <StructureSection load='1t95' size='350' side='right' scene='' caption='Shwachman-Bodian-Diamond syndrome protein [[1t95]]'> |
| - | + | ||
| - | + | ||
| - | = '''Shwachman-Bodian-Diamond | + | |
| - | + | ||
== '''Overview''' == | == '''Overview''' == | ||
| - | The human Shwachman-Bodian-Diamond syndrome(SBDS) protein belongs to a very conserved family of proteins of unknown function; orthologues found in Archaea, as well as plants and other eukaryotes <ref name="one">PMID:15701631</ref>. Evidence has been provided that the SBDS protein orthologues may play a role in RNA metabolism <ref name="one">PMID:15701631</ref>. Two groups of SBDS orthologues have been identified. Archaea, animals, and fungi have SBDS proteins with approximately 250 amino acid residues<ref name="one">PMID:15701631</ref>. However, SBDS protein of plants and protists has the C-terminal extensions around 100 to 250 amino acid residues <ref name="two">PMID: 19121363</ref>. | + | The human '''Shwachman-Bodian-Diamond syndrome (SBDS) protein''' belongs to a very conserved family of proteins of unknown function; orthologues found in Archaea, as well as plants and other eukaryotes <ref name="one">PMID:15701631</ref>. Evidence has been provided that the SBDS protein orthologues may play a role in RNA metabolism <ref name="one">PMID:15701631</ref>. Two groups of SBDS orthologues have been identified. Archaea, animals, and fungi have SBDS proteins with approximately 250 amino acid residues<ref name="one">PMID:15701631</ref>. However, SBDS protein of plants and protists has the C-terminal extensions around 100 to 250 amino acid residues <ref name="two">PMID: 19121363</ref>. |
=='''Structure'''== | =='''Structure'''== | ||
| Line 12: | Line 8: | ||
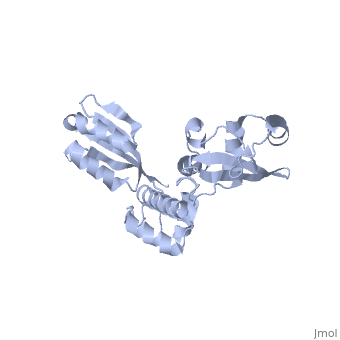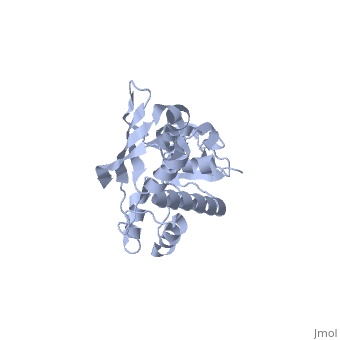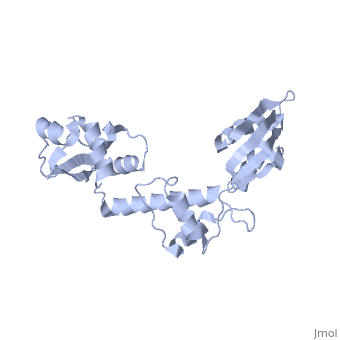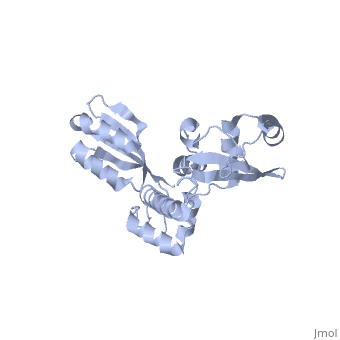
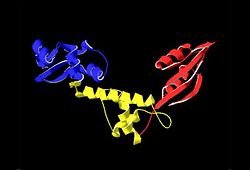
The Archaeoglobus fugidus othrologue of the human SBDS protein (AfSBDS) is a monomer of 240 amino acid residues with water molecules consisting of three domains <ref name="one">PMID:15701631</ref>. Domain I is a N-terminal domain consisting of residues 5-87. Domain II is a central domain consisting of residues 86-161. Domain III is a C-terminal domain consisting of residues 162-234 <ref name="one">PMID:15701631</ref>. This has been described as a V-shaped molecule with an overall dimension of about 54x59x73 Å (Figure. 1) <ref name="one">PMID:15701631</ref>. A central cavity (about 10 Å deep and 24 Å long) is created at the interface of the three domains <ref name="one">PMID:15701631</ref>.[[Image:SBDS 1.jpg |left|thumb|250px|'''Figure 1.''' Overall structure of the A. fulgidus SBDS orthologue described as V-shaped, ribbon representations. Domain I is coloured blue, domain II is coloured yellow, and domain III is coloured red. <ref name="one">PMID:15701631</ref>]] Domain I is a mixed αβ topology consisting of four β strands and four α helices arranged as a 3-stranded anti-parallel β sheets. Helices α1 and α2, along with the intervening loop, line the face of the cleft between Domains I and III <ref name="one">PMID:15701631</ref>. | The Archaeoglobus fugidus othrologue of the human SBDS protein (AfSBDS) is a monomer of 240 amino acid residues with water molecules consisting of three domains <ref name="one">PMID:15701631</ref>. Domain I is a N-terminal domain consisting of residues 5-87. Domain II is a central domain consisting of residues 86-161. Domain III is a C-terminal domain consisting of residues 162-234 <ref name="one">PMID:15701631</ref>. This has been described as a V-shaped molecule with an overall dimension of about 54x59x73 Å (Figure. 1) <ref name="one">PMID:15701631</ref>. A central cavity (about 10 Å deep and 24 Å long) is created at the interface of the three domains <ref name="one">PMID:15701631</ref>.[[Image:SBDS 1.jpg |left|thumb|250px|'''Figure 1.''' Overall structure of the A. fulgidus SBDS orthologue described as V-shaped, ribbon representations. Domain I is coloured blue, domain II is coloured yellow, and domain III is coloured red. <ref name="one">PMID:15701631</ref>]] Domain I is a mixed αβ topology consisting of four β strands and four α helices arranged as a 3-stranded anti-parallel β sheets. Helices α1 and α2, along with the intervening loop, line the face of the cleft between Domains I and III <ref name="one">PMID:15701631</ref>. | ||
| - | Domain I is also called the FYSH (Fungal, Yhr087wp, Shwachwan) domain because of its distinctive structural homology with a single domain protein Yhr087wp from ''S. cerevisiae'' <ref name="one">PMID:15701631</ref>. Yhr087wp is non-essential, localizing to both the nucleus and cytoplasam of yeast cells, and has a role in RNA metabolism <ref name="one">PMID:15701631</ref>. Most mutations of the SBDS protein are found to occur in this domain. Domain II is a compact three-helical bundle structurally similar to the junction recognition site of E. coli <ref name="one">PMID:15701631</ref>. Domain III is made of a four-stranded anti-parallel β-sheet with two α helices packing against the concave surface of the sheet <ref name="one">PMID:15701631</ref>. The βαββαβ folding topology is a ferredoxin-like fold which is usually seen in many RNA binding proteins, including nuclear ribonucleoproteins and small nuclear ribonucleoproteins <ref name="one">PMID:15701631</ref>. | + | Domain I is also called the FYSH (Fungal, Yhr087wp, Shwachwan) domain because of its distinctive structural homology with a single domain protein Yhr087wp from ''S. cerevisiae'' <ref name="one">PMID:15701631</ref>. Yhr087wp is non-essential, localizing to both the nucleus and cytoplasam of yeast cells, and has a role in RNA metabolism <ref name="one">PMID:15701631</ref>. Most mutations of the SBDS protein are found to occur in this domain. Domain II is a compact three-helical bundle structurally similar to the junction recognition site of ''E. coli'' <ref name="one">PMID:15701631</ref>. Domain III is made of a four-stranded anti-parallel β-sheet with two α helices packing against the concave surface of the sheet <ref name="one">PMID:15701631</ref>. The βαββαβ folding topology is a ferredoxin-like fold which is usually seen in many RNA binding proteins, including nuclear ribonucleoproteins and small nuclear ribonucleoproteins <ref name="one">PMID:15701631</ref>. |
The structure of the orthologue for the human SBDS protein from ''A. fulgidus'' (AfSBDS) has been determined because of the significant amount of amino acid conservation between the two <ref name="one">PMID:15701631</ref>. It had been hypothesized that by doing so, clues could be obtained as to what the function of the protein may be, as well as being able to determine the effects of the mutations found in SDS <ref name="one">PMID:15701631</ref>. The structure has been determined via x-ray crystallography with a resolution of 1.9Å and a free R factor of 24.9%. Crystals were grown from SeMet substituted His6-AfSBDS protein <ref name="one">PMID:15701631</ref>. This technique was an appropriate form of methodology, as it allows for structural comparison with other potentially homologous proteins, as well as allowing for the mapping of mutations onto the structure <ref name="one">PMID:15701631</ref>. | The structure of the orthologue for the human SBDS protein from ''A. fulgidus'' (AfSBDS) has been determined because of the significant amount of amino acid conservation between the two <ref name="one">PMID:15701631</ref>. It had been hypothesized that by doing so, clues could be obtained as to what the function of the protein may be, as well as being able to determine the effects of the mutations found in SDS <ref name="one">PMID:15701631</ref>. The structure has been determined via x-ray crystallography with a resolution of 1.9Å and a free R factor of 24.9%. Crystals were grown from SeMet substituted His6-AfSBDS protein <ref name="one">PMID:15701631</ref>. This technique was an appropriate form of methodology, as it allows for structural comparison with other potentially homologous proteins, as well as allowing for the mapping of mutations onto the structure <ref name="one">PMID:15701631</ref>. | ||
| Line 28: | Line 24: | ||
The yeast orthologue, Sdo1p, has been shown to participate in 60S ribosome biogenesis, as well as in the translation activation of ribosomes<ref name="two">PMID: 19121363</ref>. The C-terminal found in plants contains a zinc finger domain that can interact with nucleic acids<ref name="two">PMID: 19121363</ref>. A high binding and RNA affinity has been shown when even though the C-terminal extension of trypanosomes does not show this zinc finger domain <ref name="two">PMID: 19121363</ref>. Direct RNA binding has not been shown for orthologues without the C-terminal extention, but ribosome association has been shown<ref name="seven">PMID: 17475909</ref>. The fact that the C-terminal domain has a ferredoxin-like fold that is associated with many RNA binding proteins is also suggests of some role in RNA metabolism<ref name="one">PMID:15701631</ref>. The FYSH domain is negatively charged, and the central and C-terminal domains are mostly basic. Nucleic acid binding proteins often use basic residues to connect with the sugar phosphate backbone, further indicating an RNA link <ref name="one">PMID:15701631</ref>. | The yeast orthologue, Sdo1p, has been shown to participate in 60S ribosome biogenesis, as well as in the translation activation of ribosomes<ref name="two">PMID: 19121363</ref>. The C-terminal found in plants contains a zinc finger domain that can interact with nucleic acids<ref name="two">PMID: 19121363</ref>. A high binding and RNA affinity has been shown when even though the C-terminal extension of trypanosomes does not show this zinc finger domain <ref name="two">PMID: 19121363</ref>. Direct RNA binding has not been shown for orthologues without the C-terminal extention, but ribosome association has been shown<ref name="seven">PMID: 17475909</ref>. The fact that the C-terminal domain has a ferredoxin-like fold that is associated with many RNA binding proteins is also suggests of some role in RNA metabolism<ref name="one">PMID:15701631</ref>. The FYSH domain is negatively charged, and the central and C-terminal domains are mostly basic. Nucleic acid binding proteins often use basic residues to connect with the sugar phosphate backbone, further indicating an RNA link <ref name="one">PMID:15701631</ref>. | ||
| + | |||
| + | ==3D structures of SBDS== | ||
| + | |||
| + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | |||
| + | [[2kdo]], [[2l9n]] – SBDS – human – NMR | ||
| + | |||
| + | [[1t95]] – SBDS – ''Archaeoglobus fulgidus''<br /> | ||
| + | [[2wbm]] – SBDS homolog – ''Methanothermobacter thermautotrophicus''<br /> | ||
| + | [[5an9]], [[5anb]], [[5anc]], [[6qkl]] – SBDS in 60S ribosomal subunit – slime mold – Cryo EM<br /> | ||
=References= | =References= | ||
| + | </StructureSection> | ||
<references/> | <references/> | ||
| + | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 1.19 1.20 1.21 1.22 1.23 1.24 1.25 1.26 1.27 1.28 1.29 1.30 1.31 1.32 Shammas C, Menne TF, Hilcenko C, Michell SR, Goyenechea B, Boocock GR, Durie PR, Rommens JM, Warren AJ. Structural and mutational analysis of the SBDS protein family. Insight into the leukemia-associated Shwachman-Diamond Syndrome. J Biol Chem. 2005 May 13;280(19):19221-9. Epub 2005 Feb 8. PMID:15701631 doi:http://dx.doi.org/10.1074/jbc.M414656200
- ↑ 2.0 2.1 2.2 2.3 de Oliveira JF, Castilho BA, Sforca ML, Krieger MA, Zeri AC, Guimaraes BG, Zanchin NI. Characterization of the Trypanosoma cruzi ortholog of the SBDS protein reveals an intrinsically disordered extended C-terminal region showing RNA-interacting activity. Biochimie. 2009 Apr;91(4):475-83. Epub 2008 Dec 16. PMID:19121363 doi:10.1016/j.biochi.2008.12.001
- ↑ 3.0 3.1 3.2 3.3 3.4 Nihrane A, Sezgin G, Dsilva S, Dellorusso P, Yamamoto K, Ellis SR, Liu JM. Depletion of the Shwachman-Diamond syndrome gene product, SBDS, leads to growth inhibition and increased expression of OPG and VEGF-A. Blood Cells Mol Dis. 2009 Jan-Feb;42(1):85-91. Epub 2008 Nov 17. PMID:19014892 doi:10.1016/j.bcmd.2008.09.004
- ↑ 4.0 4.1 Rujkijyanont P, Watanabe K, Ambekar C, Wang H, Schimmer A, Beyene J, Dror Y. SBDS-deficient cells undergo accelerated apoptosis through the Fas-pathway. Haematologica. 2008 Mar;93(3):363-71. Epub 2008 Feb 11. PMID:18268284 doi:10.3324/haematol.11579
- ↑ 5.0 5.1 Austin KM, Gupta ML, Coats SA, Tulpule A, Mostoslavsky G, Balazs AB, Mulligan RC, Daley G, Pellman D, Shimamura A. Mitotic spindle destabilization and genomic instability in Shwachman-Diamond syndrome. J Clin Invest. 2008 Apr;118(4):1511-8. PMID:18324336 doi:10.1172/JCI33764
- ↑ Hesling C, Oliveira CC, Castilho BA, Zanchin NI. The Shwachman-Bodian-Diamond syndrome associated protein interacts with HsNip7 and its down-regulation affects gene expression at the transcriptional and translational levels. Exp Cell Res. 2007 Dec 10;313(20):4180-95. Epub 2007 Jul 10. PMID:17643419 doi:10.1016/j.yexcr.2007.06.024
- ↑ 7.0 7.1 7.2 7.3 7.4 Ganapathi KA, Austin KM, Lee CS, Dias A, Malsch MM, Reed R, Shimamura A. The human Shwachman-Diamond syndrome protein, SBDS, associates with ribosomal RNA. Blood. 2007 Sep 1;110(5):1458-65. Epub 2007 May 2. PMID:17475909 doi:10.1182/blood-2007-02-075184
- ↑ Savchenko A, Krogan N, Cort JR, Evdokimova E, Lew JM, Yee AA, Sanchez-Pulido L, Andrade MA, Bochkarev A, Watson JD, Kennedy MA, Greenblatt J, Hughes T, Arrowsmith CH, Rommens JM, Edwards AM. The Shwachman-Bodian-Diamond syndrome protein family is involved in RNA metabolism. J Biol Chem. 2005 May 13;280(19):19213-20. Epub 2005 Feb 8. PMID:15701634 doi:10.1074/jbc.M414421200
Proteopedia Page Contributors and Editors (what is this?)
Simmi Parhar, Jaime Prilusky, Michal Harel, Alexander Berchansky, David Canner, Andrea Gorrell