1t2z
From Proteopedia
(Difference between revisions)
| (8 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
{{Theoretical_model}} | {{Theoretical_model}} | ||
| - | {{Seed}} | ||
| - | [[Image:1t2z.png|left|200px]] | ||
| - | < | + | ==HUMAN MD-2 MODEL== |
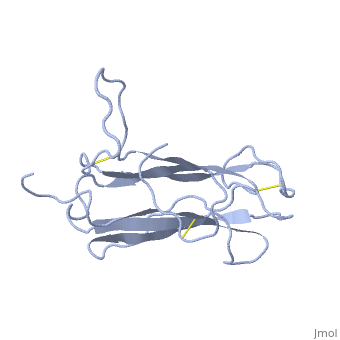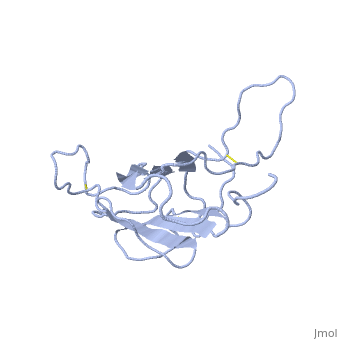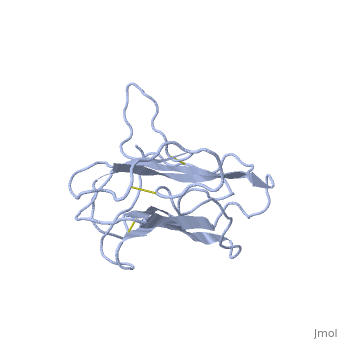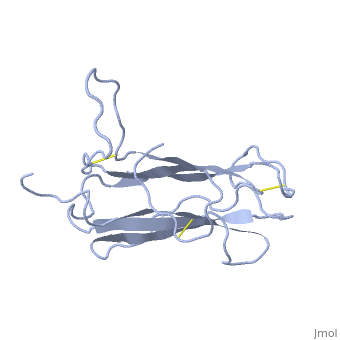
| - | The | + | <StructureSection load='1t2z' size='340' side='right'caption='[[1t2z]]' scene=''> |
| - | + | == Structural highlights == | |
| - | or the | + | <table><tr><td colspan='2'>For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1T2Z FirstGlance]. <br> |
| - | + | </td></tr><tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1t2z FirstGlance], [https://www.ebi.ac.uk/pdbsum/1t2z PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1t2z ProSAT]</span></td></tr> | |
| - | -- | + | </table> |
| - | + | <div style="background-color:#fffaf0;"> | |
| + | == Publication Abstract from PubMed == | ||
| + | The receptor complex resulting from association of MD-2 and the ectodomain of Toll-like receptor 4 (TLR4) mediates lipopolysaccharide (LPS) signal transduction across the cell membrane. We prepared a tertiary structure model of MD-2, based on the known structures of homologous lipid-binding proteins. Analysis of circular dichroic spectra of purified bacterially expressed MD-2 indicates high content of beta-type secondary structure, in agreement with the structural model. Bacterially expressed MD-2 was able to confer LPS responsiveness to cells expressing TLR4 despite lacking glycosylation. We identified several clusters of basic residues on the surface of MD-2. Mutation of each of two clusters encompassing the residues Lys(89)-Arg(90)-Lys(91) and Lys(125)-Lys(125) significantly decreased the signal transduction of the respective MD-2 mutants either upon co-expression with TLR4 or upon addition as soluble protein into the supernatant of cells overexpressing TLR4. These basic clusters lie at the edge of the beta-sheet sandwich, which in cholesterol-binding protein connected to Niemann-Pick disease C2 (NPC2), dust mite allergen Der p2, and ganglioside GM2-activator protein form a hydrophobic pocket. In contrast, mutation of another basic cluster composed of Arg(69)-Lys(72), which according to the model lies further apart from the hydrophobic pocket only weakly decreased MD-2 activity. Furthermore, addition of the peptide, comprising the surface loop between Cys(95) and Cys(105), predicted by model, particularly in oxidized form, decreased LPS-induced production of tumor necrosis factor alpha and interleukin-8 upon application to monocytic cells and fibroblasts, respectively, supporting its involvement in LPS signaling. Our structural model of MD-2 is corroborated by biochemical analysis and contributes to the unraveling of molecular interactions in LPS recognition. | ||
| - | + | Structural model of MD-2 and functional role of its basic amino acid clusters involved in cellular lipopolysaccharide recognition.,Gruber A, Mancek M, Wagner H, Kirschning CJ, Jerala R J Biol Chem. 2004 Jul 2;279(27):28475-82. Epub 2004 Apr 24. PMID:15111623<ref>PMID:15111623</ref> | |
| - | + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |
| - | + | </div> | |
| - | + | <div class="pdbe-citations 1t2z" style="background-color:#fffaf0;"></div> | |
| - | + | == References == | |
| - | + | <references/> | |
| - | + | __TOC__ | |
| - | + | </StructureSection> | |
| - | == | + | [[Category: Theoretical Model]] |
| - | + | [[Category: Large Structures]] | |
| - | + | ||
| - | == | + | |
| - | < | + | |
[[Category: Gruber, A]] | [[Category: Gruber, A]] | ||
[[Category: Jerala, R]] | [[Category: Jerala, R]] | ||
| Line 30: | Line 27: | ||
[[Category: Mancek, M]] | [[Category: Mancek, M]] | ||
[[Category: Wagner, H]] | [[Category: Wagner, H]] | ||
| - | |||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu Apr 8 08:58:55 2010'' | ||
Current revision
| Theoretical Model: The protein structure described on this page was determined theoretically, and hence should be interpreted with caution. |
HUMAN MD-2 MODEL
| |||||||||||