User:Tsung-Yi Lin/Sandbox 1
From Proteopedia
| (14 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | [[Image:intactModelLargeText.jpg|frame|Bacterial chemotaxis receptor]] | ||
| - | |||
A [[CBI Molecule]] being studied in the [http://www.umass.edu/cbi/ University of Massachusetts Amherst Chemistry-Biology Interface Program] at UMass Amherst and on display at the [http://www.molecularplayground.org/ Molecular Playground]. | A [[CBI Molecule]] being studied in the [http://www.umass.edu/cbi/ University of Massachusetts Amherst Chemistry-Biology Interface Program] at UMass Amherst and on display at the [http://www.molecularplayground.org/ Molecular Playground]. | ||
| - | Many bacteria can "smell" their surroundings and "choose" where to go. They detect molecules such as amino acids or sugars using receptors that bind these molecules and transmit a signal into the cell. This signal controls several proteins which ultimately control the direction of rotation of the motors that rotate the flagella. One direction causes the cell to continue swimming; the other direction causes the cell to tumble. When an attractant molecule binds, the receptor signals: "Things look good, keep swimming!" The opposite signal occurs when bacteria sense a repellant or less attractant molecules: "Time to tumble and try a new swimming direction." | ||
| + | Polyketides are a large class of natural products produced by bacteria, fungi, and plants. Polyketides exhibit diverse structures and a wide variety of biological activities, which includes antibiotic, antitumor, anticancer, and other biological properties. | ||
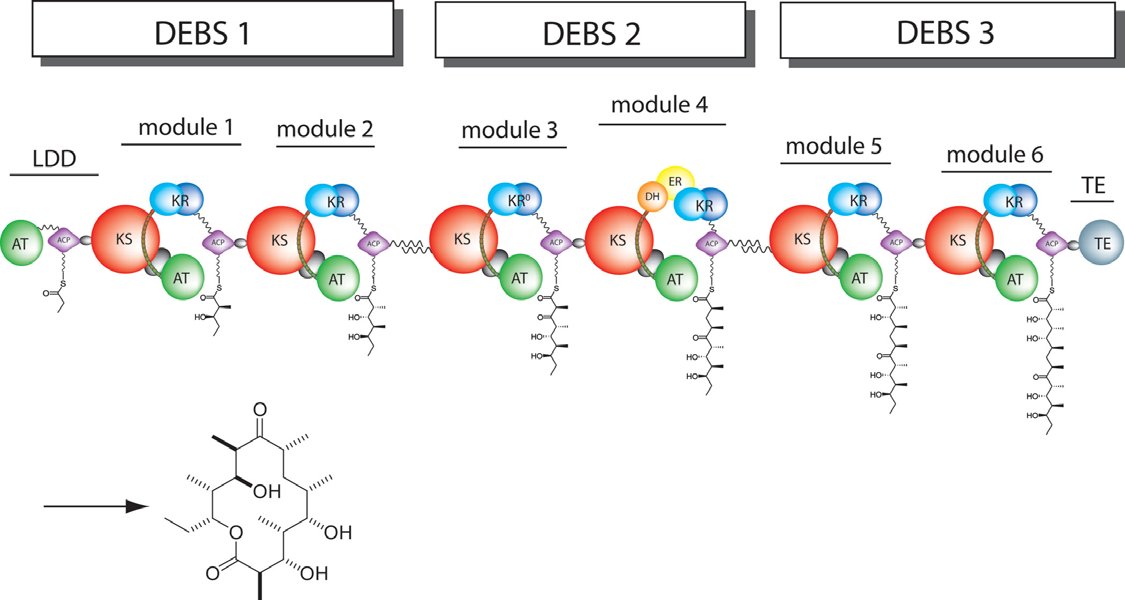
| - | A | + | In nature, polyketides are synthesized by large multifunctional enzymes called polyketide synthases. Among several characterized PKSs, the biosynthesis of the polyketide Core of erythromycin A, 6-deoxyerythronolide B (6-dEB), has provided the paradigm for understanding the structure and function of the PKSs that are responsible for assembling complex polyketides . |
| + | |||
| + | [[Image:DEBS.png|frame|6-deoxyerythronolide B synthase (DEBS)|left|300px]] | ||
| + | The 6-deoxyerythronolide B synthase (DEBS), which catalyzes the formation of 6-dEB, consists of three large polypeptides, DEBS1, DEBS2 and DEBS3, each 330 to 370 kD in size, and each polypeptide in DEBS is composed of two modules, where a module includes all the catalytic domains responsible for one round of chain extension and modification on the growing polyketide intermediate. Each module minimally contains three essential domains; a β-keto acyl synthase, simply referred to as ketosynthase (KS), an acyl transferase (AT) and an acyl carrier protein (ACP). All of them catalyze extender units adding to the growing polyketide chain. In detail, AT domain selects the appropriate monomer unit and transfers the carbon extender units from acyl-CoA metabolite onto the phosphopantetheinyl arm of ACP, and KS domains catalyzes decarboxylative condensation between the growing polyketide chain from the previous module and an ACP-bound extender unit. | ||
| + | |||
| + | After the extender unit was added onto the elongating polyketide chain, it can be further processed by optional tailoring domains, including ketoreductases (KRs), dehydratases (DHs), and enoyl reductases (ERs), to change the oxidation state of each ketide-unit and to yield a hydroxyl, enoyl, or methylene group that may be present in the final product. In addition, the thioesterase (TE) domain that located at the C-terminus of module 6 of DEBS catalyzes the macrocyclization and releases the final product 6-dEB. | ||
| + | |||
| + | |||
| + | ===KS-AT=== | ||
| + | |||
| + | <applet load='2HG4' size='[450,338]' frame='true' align='right' | ||
| + | caption='Structure of the ketosynthase-acyltransferase didomain of module 5 from DEBS. (2HG4)' scene='User:Tsung-Yi Lin/Sandbox_1/> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ===ACP=== | ||
| + | |||
| + | <applet load='2JU1' size='[450,338]' frame='true' align='right' | ||
| + | caption='Solution structure of acyl carrier protein domain from module 2 of 6-deoxyerythronolide B synthase (DEBS) (2JU1)' scene='User:Tsung-Yi Lin/Sandbox_1/> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ===DH=== | ||
| - | {{Clear}} | ||
<applet load='3EL6' size='[450,338]' frame='true' align='right' | <applet load='3EL6' size='[450,338]' frame='true' align='right' | ||
| - | caption='Crystal Structure of the Erythromycin Dehydratase (3EL6)'> | + | caption='Crystal Structure of the Erythromycin Dehydratase (3EL6)' scene='User:Tsung-Yi Lin/Sandbox_1/> |
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ===TE=== | ||
| + | |||
| + | <applet load='1MO2' size='[450,338]' frame='true' align='right' | ||
| + | caption='Thioesterase Domain from 6-Deoxyerythronolide Synthase (DEBS TE)(1MO2)' scene='User:Tsung-Yi Lin/Sandbox_1/> | ||
Current revision
A CBI Molecule being studied in the University of Massachusetts Amherst Chemistry-Biology Interface Program at UMass Amherst and on display at the Molecular Playground.
Polyketides are a large class of natural products produced by bacteria, fungi, and plants. Polyketides exhibit diverse structures and a wide variety of biological activities, which includes antibiotic, antitumor, anticancer, and other biological properties.
In nature, polyketides are synthesized by large multifunctional enzymes called polyketide synthases. Among several characterized PKSs, the biosynthesis of the polyketide Core of erythromycin A, 6-deoxyerythronolide B (6-dEB), has provided the paradigm for understanding the structure and function of the PKSs that are responsible for assembling complex polyketides .
The 6-deoxyerythronolide B synthase (DEBS), which catalyzes the formation of 6-dEB, consists of three large polypeptides, DEBS1, DEBS2 and DEBS3, each 330 to 370 kD in size, and each polypeptide in DEBS is composed of two modules, where a module includes all the catalytic domains responsible for one round of chain extension and modification on the growing polyketide intermediate. Each module minimally contains three essential domains; a β-keto acyl synthase, simply referred to as ketosynthase (KS), an acyl transferase (AT) and an acyl carrier protein (ACP). All of them catalyze extender units adding to the growing polyketide chain. In detail, AT domain selects the appropriate monomer unit and transfers the carbon extender units from acyl-CoA metabolite onto the phosphopantetheinyl arm of ACP, and KS domains catalyzes decarboxylative condensation between the growing polyketide chain from the previous module and an ACP-bound extender unit.
After the extender unit was added onto the elongating polyketide chain, it can be further processed by optional tailoring domains, including ketoreductases (KRs), dehydratases (DHs), and enoyl reductases (ERs), to change the oxidation state of each ketide-unit and to yield a hydroxyl, enoyl, or methylene group that may be present in the final product. In addition, the thioesterase (TE) domain that located at the C-terminus of module 6 of DEBS catalyzes the macrocyclization and releases the final product 6-dEB.
Contents |
KS-AT
|
ACP
|
DH
|
TE
|