This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
3m9s
From Proteopedia
(Difference between revisions)
| (8 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | {{Seed}} | ||
| - | [[Image:3m9s.jpg|left|200px]] | ||
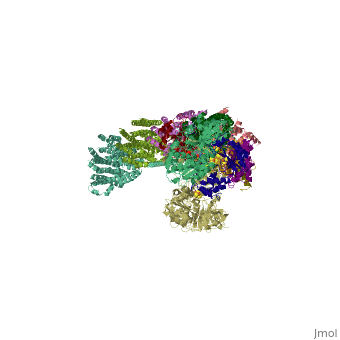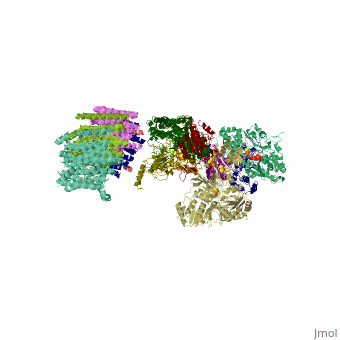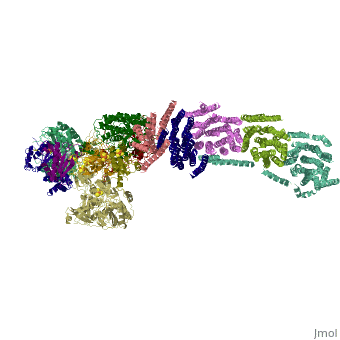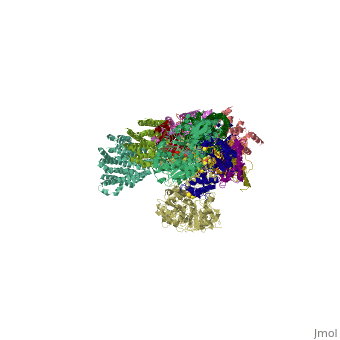
| - | < | + | ==Crystal structure of respiratory complex I from Thermus thermophilus== |
| - | The | + | <StructureSection load='3m9s' size='340' side='right'caption='[[3m9s]], [[Resolution|resolution]] 4.50Å' scene=''> |
| - | + | == Structural highlights == | |
| - | + | <table><tr><td colspan='2'>[[3m9s]] is a 20 chain structure with sequence from [https://en.wikipedia.org/wiki/Thermus_thermophilus_HB8 Thermus thermophilus HB8]. The December 2011 RCSB PDB [https://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month] feature on ''Complex I'' by David Goodsell is [https://dx.doi.org/10.2210/rcsb_pdb/mom_2011_12 10.2210/rcsb_pdb/mom_2011_12]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3M9S OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=3M9S FirstGlance]. <br> | |
| - | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 4.5Å</td></tr> | |
| - | + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=FES:FE2/S2+(INORGANIC)+CLUSTER'>FES</scene>, <scene name='pdbligand=FMN:FLAVIN+MONONUCLEOTIDE'>FMN</scene>, <scene name='pdbligand=SF4:IRON/SULFUR+CLUSTER'>SF4</scene></td></tr> | |
| - | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=3m9s FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3m9s OCA], [https://pdbe.org/3m9s PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=3m9s RCSB], [https://www.ebi.ac.uk/pdbsum/3m9s PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=3m9s ProSAT]</span></td></tr> | |
| + | </table> | ||
| + | == Function == | ||
| + | [https://www.uniprot.org/uniprot/NQO9_THET8 NQO9_THET8] NDH-1 shuttles electrons from NADH, via FMN and iron-sulfur (Fe-S) centers, to quinones in the respiratory chain. The immediate electron acceptor for the enzyme in this species is menaquinone. Couples the redox reaction to proton translocation (for every two electrons transferred, four hydrogen ions are translocated across the cytoplasmic membrane), and thus conserves the redox energy in a proton gradient required for the synthesis of ATP. The role of the nqo9 subunit appears to provide a 'connecting chain' of two clusters between cluster N5 and the terminal cluster N2, and to stabilize the structure of the complex by interacting with other subunits.[HAMAP-Rule:MF_01351] | ||
| + | == Evolutionary Conservation == | ||
| + | [[Image:Consurf_key_small.gif|200px|right]] | ||
| + | Check<jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/m9/3m9s_consurf.spt"</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | ||
| + | <text>to colour the structure by Evolutionary Conservation</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=3m9s ConSurf]. | ||
| + | <div style="clear:both"></div> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
| + | Complex I is the first enzyme of the respiratory chain and has a central role in cellular energy production, coupling electron transfer between NADH and quinone to proton translocation by an unknown mechanism. Dysfunction of complex I has been implicated in many human neurodegenerative diseases. We have determined the structure of its hydrophilic domain previously. Here, we report the alpha-helical structure of the membrane domain of complex I from Escherichia coli at 3.9 A resolution. The antiporter-like subunits NuoL/M/N each contain 14 conserved transmembrane (TM) helices. Two of them are discontinuous, as in some transporters. Unexpectedly, subunit NuoL also contains a 110-A long amphipathic alpha-helix, spanning almost the entire length of the domain. Furthermore, we have determined the structure of the entire complex I from Thermus thermophilus at 4.5 A resolution. The L-shaped assembly consists of the alpha-helical model for the membrane domain, with 63 TM helices, and the known structure of the hydrophilic domain. The architecture of the complex provides strong clues about the coupling mechanism: the conformational changes at the interface of the two main domains may drive the long amphipathic alpha-helix of NuoL in a piston-like motion, tilting nearby discontinuous TM helices, resulting in proton translocation. | ||
| - | + | The architecture of respiratory complex I.,Efremov RG, Baradaran R, Sazanov LA Nature. 2010 May 27;465(7297):441-5. PMID:20505720<ref>PMID:20505720</ref> | |
| + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| + | </div> | ||
| + | <div class="pdbe-citations 3m9s" style="background-color:#fffaf0;"></div> | ||
| - | == | + | ==See Also== |
| - | + | *[[NADH-quinone oxidoreductase|NADH-quinone oxidoreductase]] | |
| - | + | == References == | |
| - | + | <references/> | |
| - | [[Category: | + | __TOC__ |
| - | [[Category: | + | </StructureSection> |
| - | [[Category: | + | [[Category: Complex I]] |
| - | [[Category: | + | [[Category: Large Structures]] |
| - | [[Category: | + | [[Category: RCSB PDB Molecule of the Month]] |
| - | [[Category: | + | [[Category: Thermus thermophilus HB8]] |
| - | [[Category: | + | [[Category: Baradaran R]] |
| - | + | [[Category: Efremov RG]] | |
| - | + | [[Category: Sazanov LA]] | |
Current revision
Crystal structure of respiratory complex I from Thermus thermophilus
| |||||||||||