We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Ferredoxin
From Proteopedia
(Difference between revisions)
| (59 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
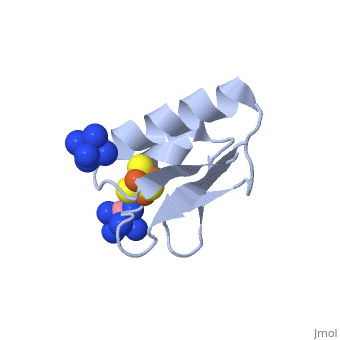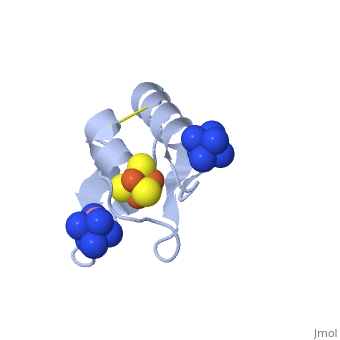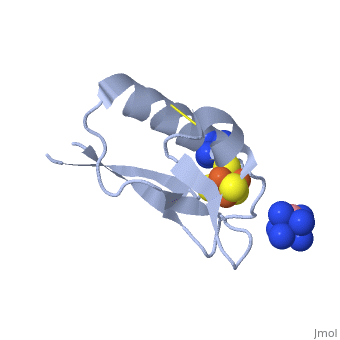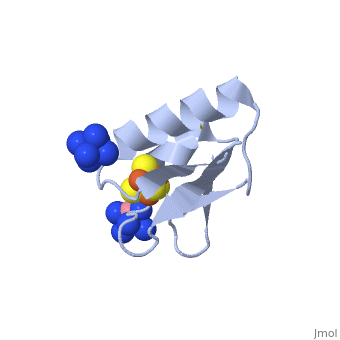
| - | + | <StructureSection load='2z8q' size='450' side='right' scene='' caption='Ferredoxin with Fe4S4 cluster complex with cobalt hexamine (PDB code [[2z8q]])'> | |
| - | < | + | |
| - | ''' | + | |
| - | + | ==Function== | |
| - | + | ||
| + | [[Ferredoxin]] (Fd) is found in chloroplasts which mediates electron transfer and contains an iron-sulfur cluster. It is involved in the photosynthesis process where its iron atoms accept or discharge electrons when they are being oxidized or reduced. The iron-sulfur cluster can contain 2Fe-2S and is termed plant-like or 3Fe-4S or 4Fe-4S clusters. | ||
| + | *'''Adrenodoxin''' (ADR) is a ferredoxin containing a 2Fe-2S group involved in electron transfer from NADPH+ to a cytochrome P-450 in the adrenal gland<ref>PMID:22556163</ref>.. | ||
| + | *'''Putidaredoxin''' (PUT) and '''terpredoxin''' (TER) are involved in the same reaction in bacteria and contain a 2Fe-2S group<ref>PMID:10220356</ref>. | ||
| - | === | + | ==D14C variant of ''Pyrococcus furiosus'' ferredoxin<ref>DOI 10.1007/s00775-011-0778-7</ref>== |
| + | Structures of the all cysteinyl coordinated D14C variant of ferredoxin from the hyperthermophilic archaeon ''Pyrococcus furiosus'' have been determined for the <scene name='Journal:JBIC:10/Cv1/5'>[4Fe-4S]</scene> <-> and <scene name='Journal:JBIC:10/Cv1/6'>[3Fe-4S]</scene> forms (<scene name='Journal:JBIC:10/Cv1/8'>click to enlarge</scene>). The [4Fe-4S] form diffracted to 1.7 Å and two different types of molecules were found in the crystal ([[2z8q]]). They have different crystal packing and intramolecular disulfide bond conformation. The crystal packing reveals a <scene name='Journal:JBIC:10/Cv/5'>beta-sheet interaction between A molecules</scene> (shown in <font color='blue'><b>blue</b></font> and <span style="color:green;background-color:black;font-weight:bold;">green</span>) in adjacent asymmetric units, while <scene name='Journal:JBIC:10/Cv/6'>B molecules are packed as monomers in a less rigid position next to the A-A extended beta-sheet dimers</scene> (shown in <font color='red'><b>red</b></font> and <span style="color:yellow;background-color:black;font-weight:bold;">yellow</span>). The <scene name='Journal:JBIC:10/Cv1/9'>intramolecular disulfide bond in the A molecules is in a double conformation</scene> while the intramolecular disulfide bond in the <scene name='Journal:JBIC:10/Cv1/10'>B molecules is in a single conformation</scene> (<scene name='Journal:JBIC:10/Cv1/11'>click to see morph</scene>, molecule A is shown in <font color='blue'><b>blue</b></font> and molecule B in <span style="color:green;background-color:black;font-weight:bold;">green</span>). | ||
| - | [[ | + | [[Image:Schem1.png|left|300px|thumb|pH dependent equilibrium of D14C [3Fe-4S] ''P. furiosus'' ferredoxin between protonated and deprotonated monomers and formation of a disulfide bonded dimer from deprotonated monomers. Fd is short for ferredoxin.]] |
| - | + | {{Clear}} | |
| - | [ | + | Two forms of D14C [3Fe-4S] ''Pyrococcus furiosus'' ferredoxin are obtained when purified at pH 8.0: a monomer and a dimer connected by an intermolecular disulfide bond (see static image above). When purified at pH 5.8, only the monomer is obtained. The [3Fe-4S] form diffracted to 2.8 Å resolution and showed only the <scene name='Journal:JBIC:10/Cv1/13'>monomeric form, which resembles molecule A of D14C [4Fe-4S] Pyrococcus furiosus ferredoxin</scene>. Crystal packing in <scene name='Journal:JBIC:10/Cv2/7'>D14C [3Fe-4S] ferredoxin is as extended beta-sheet dimers of adjacent molecules</scene> (shown in <font color='red'><b>red</b></font> and <font color='orange'><b>orange</b></font>), which is the same as <scene name='Journal:JBIC:10/Cv2/9'>WT [3Fe-4S] P. furiosus ferredoxin</scene> ([[1sj1]], shown in <font color='blue'><b>blue</b></font> and <font color='cyan'><b>cyan</b></font>) even though the space groups are different (see also corresponding side views for <scene name='Journal:JBIC:10/Cv2/8'>D14C [3Fe-4S]</scene>) and <scene name='Journal:JBIC:10/Cv2/10'>WT [3Fe-4S]</scene>). |
| - | + | ||
| - | [ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | [ | + | |
| - | [[ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | [ | + | |
| - | + | ||
| - | [ | + | |
| - | == | + | ==ISC-like [2Fe-2S] ferredoxin (FdxB) dimer from ''Pseudomonas putida'' JCM 20004<ref>DOI:10.1007/s00775-011-0793-8</ref>== |
| + | Biological iron-sulfur (Fe-S) clusters are functionally versatile, modular prosthetic groups. The electronic structure and the site of iron reduction of these protein-bound cofactors account for the electron transfer function and mechanism. In the present work we have solved the structure of the ISC-like [2Fe-2S] ferredoxin called FdxB from the non-pathogenic gammaproteobacterium ''Pseudomonas putida'' JCM 20004 (formerly ''Pseudomonas ovalis'' IAM 1002) ([[3ah7]]). This FdxB protein contains an adrenodoxin (Adx) like, redox-active [2Fe-2S] cluster, which plays an essential role in the de novo iron-sulfur cluster assembly (ISC) system. It is encoded by the fdxB gene as a constituent of the cognate iscR-iscS1-iscU-iscA-hscB-hscA-fdxB gene cluster for the ISC system (DDBJ-EMBL-GenBank code AB109467). In ''P. putida'' the ISC pathway is apparently the sole system for ''in vivo'' Fe-S cluster assembly whereas the SUF pathway is missing in the bacterial genome (unlike in ''Escherichia coli''). | ||
| - | + | The <scene name='Journal:JBIC:12/Cv1/1'>FdxB structure</scene> has a βαββαβ fold with the β-grasp/ubiquitin-like fold motif as found in regular eukaryal and bacterial [2Fe-2S] ferredoxins (e.g. [[1i7h]], [[1cje]], [[1e9m]]). FdxB is folded into an (α+β) <scene name='Journal:JBIC:12/Cv1/2'>core fold domain and an extended C-terminal tail</scene>. In the lattice <scene name='Journal:JBIC:12/Cv1/3'>FdxB was found to be homo-dimeric, </scene> displaying the <scene name='Journal:JBIC:12/Cv1/13'>isologous association of the extended C-terminal tail from each protomer</scene>. Each protomer binds a <scene name='Journal:JBIC:12/Cv1/4'>[2Fe-2S] cluster</scene> that is <scene name='Journal:JBIC:12/Cv1/5'>coordinated by four terminal cysteine sulfur atoms</scene>, where the <scene name='Journal:JBIC:12/Cv1/7'>outermost iron (Fe1) near the protein surface is coordinated by Cys41S and Cys47S</scene> and the <scene name='Journal:JBIC:12/Cv1/8'>innermost iron (Fe2) by Cys50S and Cys86S</scene>. In the <scene name='Journal:JBIC:12/Cv1/9'>dimeric structure, two [2Fe-2S] clusters are separated at the closest iron-to-iron (Fe1-Fe1) distance of 25 A</scene>, suggesting that a rapid interprotomer electron transfer between them would be unlikely to occur. In the place of the consensus free cysteine usually present near the [2Fe-2S] cluster of ISC-like ferredoxins, FdxB has the <scene name='Journal:JBIC:12/Cv1/10'>Lys45 side chain which forms a salt-bridge interaction with Asp65</scene> Oδ2. Thus, the overall FdxB structural features argue for its primarily electron transfer role in the cognate ISC system, rather than the direct catalytic function. | |
| - | + | ||
| - | + | ||
| - | [ | + | |
| - | [[ | + | |
| - | [[ | + | |
| - | + | ||
| - | [ | + | |
| - | [ | + | |
| - | [ | + | |
| - | + | With the molecular structural frame determined from the FdxB structure, our electron-nuclear double resonance (ENDOR) analysis has allowed to determine the average g<sub>max</sub> direction of the reduced FdxB, which is skewed, pointing roughly towards Cys50 Cα and forming an angle of about 27.3 (±4) degrees with the normal of the [2Fe-2S] plane, while the g<sub>int</sub>- and g<sub>min</sub>-directions are distributed in a plane tilted toward the cluster plane (see image below). | |
| + | [[Image:FdxBFig8.jpg|left|400px|thumb|Skewed orientations of the g<sub>max</sub> component (red) with respect to | ||
| + | the molecular frame of the [2Fe–2S] cluster of FdxB.]] | ||
| + | {{Clear}} | ||
| + | The site of reduced iron in the reduced FdxB is the outermost Fe1 site with the low negative spin density, while the innermost Fe2 site with the high positive spin population is the non-reducible iron retaining the Fe3+-valence of a reduced cluster. From a structural point of view, the larger number of polarized (or polarizable) bonds (NH, OH) and the <scene name='Journal:JBIC:12/Cv1/15'>extended hydrogen bonding network around Fe1 in FdxB may be the crucial factor favoring the accommodation of the reducing electron at the outermost Fe1 site</scene>. These results suggest a significant distortion of the electronic structure of the reduced [2Fe-2S] cluster under the influence of the protein environment around each iron site in general. | ||
| - | [ | + | == Heterometallic [AgFe<sub>3</sub>S<sub>4</sub>] ferredoxin variants – synthesis, characterization and the first crystal structure of an engineered heterometallic iron-sulfur protein <ref >pmid 23296387 </ref>== |
| - | [[ | + | The crystal structure of the ''Pyrococcus furiosus'' (Pf) ferredoxin (Fd) D14C variant with the novel [AgFe<sub>3</sub>S<sub>4</sub>] heterometallic cluster was determined to 1.95 Å resolution (PBD entry [[4dhv]]), being the first reported structure of an engineered heterometallic iron-sulfur protein. |
| - | [[ | + | The crystal structure of the <scene name='Journal:JBIC:19/Cv/4'>monomeric form</scene> shows that the <scene name='Journal:JBIC:19/Cv/6'>silver (I) ion is part of the cluster</scene> (clearly seen on the electron density map), as predicted from previous spectroscopic and electrochemical studies. The heterometal is coordinated to the <scene name='Journal:JBIC:19/Cv/5'>three inorganic sulfides of the cluster</scene> and to the <scene name='Journal:JBIC:19/Cv/7'>thiolate group of residue 14</scene> (residues <scene name='Journal:JBIC:19/Cv/8'>Cys11, Cys17 and Cys56</scene> are coordinated with Fe ions of heterometal), <scene name='Journal:JBIC:19/Cv/9'>replacing the aboriginal Fe ion in the all-iron coordinated</scene> [Fe<sub>4</sub>S<sub>4</sub>] D14C variant ([[2z8q]], <span style="color:cyan;background-color:black;font-weight:bold;">heterometallic [AgFe<sub>3</sub>S<sub>4</sub>] protein is in cyan</span> and <span style="color:lime;background-color:black;font-weight:bold;">homometallic [Fe<sub>4</sub>S<sub>4</sub>] is in green</span>) and completing the incomplete cuboidal cluster present in the [Fe<sub>3</sub>S<sub>4</sub>] WT Pf Fd (PDB: [[1sj1]]) and its D14C (PDB: [[3pni]]) variant (for more details see also [http://proteopedia.org/w/Journal:JBIC:10 "Crystal structures of the all cysteinyl coordinated D14C variant of ''Pyrococcus furiosus'' ferredoxin: (4Fe-4S) <-> (3Fe-4S) cluster conversion"]). Structure alignment of backbone atoms from the heterometallic [AgFe<sub>3</sub>S<sub>4</sub>] protein and the homometallic [Fe<sub>4</sub>S<sub>4</sub>] D14C variant ([[2z8q]]) shows <scene name='Journal:JBIC:19/Cv/10'>very minor differences</scene>, i.e. the root mean square deviation (RMSD) is 0.4 – 0.7 Å, observed due to the alternate conformation of the main chain atoms, flexible loops and small changes at the N- and C-termini (<span style="color:cyan;background-color:black;font-weight:bold;">heterometallic [AgFe<sub>3</sub>S<sub>4</sub>] protein is in cyan</span> and <span style="color:lime;background-color:black;font-weight:bold;">homometallic [Fe<sub>4</sub>S<sub>4</sub>] is in green</span>). <scene name='Journal:JBIC:19/Cv/14'>More significant difference</scene> can be seen in the superimposed Fe–S clusters (atom colors corresponding to <span style="color:yellow;background-color:black;font-weight:bold;">yellow: S</span>, <span style="color:orange;background-color:black;font-weight:bold;">orange: Fe</span>, <span style="color:gray;background-color:black;font-weight:bold;">gray: Ag</span>) from these two variants, which is due to the presence of the second row transition metal ion (Ag) coordinated to the four S-ligands, i.e. the presence of Ag results in a distorted geometry of the cluster compared to the all-iron arrangement, <scene name='Journal:JBIC:19/Cv/12'>because to the longer</scene> Ag – S bond lengths compared to Fe – S bonds. However, the S – Ag – S <scene name='Journal:JBIC:19/Cv/13'>bond angles</scene> are still close to the expected 90° for a primitive cubic system. |
| - | [[ | + | |
| - | [[ | + | |
| - | [ | + | |
| - | == | + | ==3D structures of ferredoxin== |
| + | [[Ferredoxin 3D structures]] | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | ==References== |
| + | <references/> | ||
| - | + | [[Category:Topic Page]] | |
| - | + | [[Category:Iron]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | [[Category: | + | |
| - | [[Category: | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
| |||||||||||
References
- ↑ Ewen KM, Ringle M, Bernhardt R. Adrenodoxin--a versatile ferredoxin. IUBMB Life. 2012 Jun;64(6):506-12. PMID:22556163 doi:10.1002/iub.1029
- ↑ Mo H, Pochapsky SS, Pochapsky TC. A model for the solution structure of oxidized terpredoxin, a Fe2S2 ferredoxin from Pseudomonas. Biochemistry. 1999 Apr 27;38(17):5666-75. PMID:10220356 doi:http://dx.doi.org/10.1021/bi983063r
- ↑ Lovgreen MN, Martic M, Windahl MS, Christensen HE, Harris P. Crystal structures of the all-cysteinyl-coordinated D14C variant of Pyrococcus furiosus ferredoxin: [4Fe-4S] <--> [3Fe-4S] cluster conversion. J Biol Inorg Chem. 2011 Apr 12. PMID:21484348 doi:10.1007/s00775-011-0778-7
- ↑ Iwasaki T, Kappl R, Bracic G, Shimizu N, Ohmori D, Kumasaka T. ISC-like [2Fe-2S] ferredoxin (FdxB) dimer from Pseudomonas putida JCM 20004: structural and electron-nuclear double resonance characterization. J Biol Inorg Chem. 2011 Jun 7. PMID:21647778 doi:10.1007/s00775-011-0793-8
- ↑ Martic M, Jakab-Simon IN, Haahr LT, Hagen WR, Christensen HE. Heterometallic [AgFe(3)S (4)] ferredoxin variants: synthesis, characterization, and the first crystal structure of an engineered heterometallic iron-sulfur protein. J Biol Inorg Chem. 2013 Feb;18(2):261-76. doi: 10.1007/s00775-012-0971-3. Epub, 2013 Jan 8. PMID:23296387 doi:10.1007/s00775-012-0971-3
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky, Eran Hodis, Wayne Decatur, David Canner
![pH dependent equilibrium of D14C [3Fe-4S] P. furiosus ferredoxin between protonated and deprotonated monomers and formation of a disulfide bonded dimer from deprotonated monomers. Fd is short for ferredoxin.](/wiki/images/thumb/0/0b/Schem1.png/300px-Schem1.png)
![Skewed orientations of the gmax component (red) with respect to the molecular frame of the [2Fe–2S] cluster of FdxB.](/wiki/images/2/29/FdxBFig8.jpg)
