Sandbox Erice 8
From Proteopedia
| (7 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| + | [[Image:3cl0_8.png|thumb|left|150px|influenza neuraminidase (3cl0)]] | ||
| + | |||
'''Influenza neuraminidase''' is a glycoprotein in the influenza virus membrane. Before an infected cell can release the virus into its surroundings to infect new cells, neuraminidase must cleave sialic acid from both virus and cellular glycoproteins. Neuraminidase is a homotetramer -- here we will examine only one monomer. | '''Influenza neuraminidase''' is a glycoprotein in the influenza virus membrane. Before an infected cell can release the virus into its surroundings to infect new cells, neuraminidase must cleave sialic acid from both virus and cellular glycoproteins. Neuraminidase is a homotetramer -- here we will examine only one monomer. | ||
== Exploring the Structure == | == Exploring the Structure == | ||
| + | <applet load='3cl0' size='300' frame='true' align='right' caption='Insert caption here' /> | ||
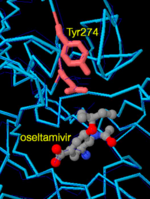
| - | Because of its important role in virus infectivity, several anti-viral drugs have been designed to target neuraminidase, including oseltamivir (Tamiflu) and zanamivir (Relenza). Oseltamavir binding to neuraminidase moves glutamate 276 towards histidine 274, making more room for oseltamavir to bind tightly (PDB entry [[2hu4]]). But, in a common mutant (H274Y), a larger tyrosine replaces the smaller histidine 274, preventing glutamate 276 from moving to make room for oseltamavir binding, resulting in weaker drug binding and thus resistance (PDB entry [[3cl0]]). Luckily the H274Y neuraminidase mutant is still susceptible to zanamivir, which is smaller than oseltamavir. | + | Because of its important role in virus infectivity, several anti-viral drugs have been designed to target neuraminidase, including <scene name='Sandbox_Erice_8/Zoom_3cl0/1'>oseltamivir</scene> (Tamiflu) and zanamivir (Relenza). Oseltamavir binding to neuraminidase moves glutamate 276 towards histidine 274, making more room for oseltamavir to bind tightly (PDB entry [[2hu4]]). But, in a common mutant (<scene name='Sandbox_Erice_8/Oseltamivir/1'>H274Y</scene>), a larger tyrosine replaces the smaller histidine 274, preventing glutamate 276 from moving to make room for oseltamavir binding, resulting in weaker drug binding and thus resistance (PDB entry [[3cl0]]). Luckily the H274Y neuraminidase mutant is still susceptible to zanamivir, which is smaller than oseltamavir. |
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
Influenza neuraminidase is a glycoprotein in the influenza virus membrane. Before an infected cell can release the virus into its surroundings to infect new cells, neuraminidase must cleave sialic acid from both virus and cellular glycoproteins. Neuraminidase is a homotetramer -- here we will examine only one monomer.
Exploring the Structure
|
Because of its important role in virus infectivity, several anti-viral drugs have been designed to target neuraminidase, including (Tamiflu) and zanamivir (Relenza). Oseltamavir binding to neuraminidase moves glutamate 276 towards histidine 274, making more room for oseltamavir to bind tightly (PDB entry 2hu4). But, in a common mutant (), a larger tyrosine replaces the smaller histidine 274, preventing glutamate 276 from moving to make room for oseltamavir binding, resulting in weaker drug binding and thus resistance (PDB entry 3cl0). Luckily the H274Y neuraminidase mutant is still susceptible to zanamivir, which is smaller than oseltamavir.