Dinoflagellate luciferase
From Proteopedia
(Difference between revisions)
m (→Related Links) |
|||
| (17 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
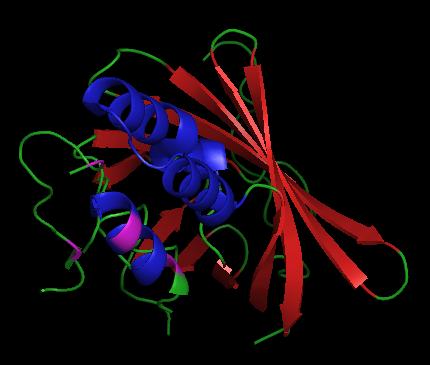
| - | + | <StructureSection load='1vpr' size='300' side='right' scene='' caption='Luciferase domain from a Dinoflagellate. Selenomethionines shown as space-filling objects [[1vpr]]'> | |
| - | + | ||
| - | + | ||
| - | + | ||
== Introduction == | == Introduction == | ||
| - | ''Lingulodinium polyedrum'', a marine dinoflagellate often responsible for red tide, | + | ''Lingulodinium polyedrum'', a marine dinoflagellate often responsible for red tide, possesses a unique [[luciferase]] enyzme. When mechanically stimulated, the organism uses this enzyme to produce a blue light, likely for use in quorum sensing. Other luciferase enzymes typically produce green-yellow to red light. Also, while all luciferase enzymes produce light through oxidation of luciferin, the biochemical mechanism by which this is achieved varies. This means that while the result is the same, there is low similarity to bacterial or firefly luciferases. |
In ''L. polyedrum'', the luciferase enzyme is a single polypeptide chain folded into 3 similar domains. Interestingly, all three domains appear to be distinct luciferase centres with their own catalytic activities <ref name="main">PMID:15665092</ref> | In ''L. polyedrum'', the luciferase enzyme is a single polypeptide chain folded into 3 similar domains. Interestingly, all three domains appear to be distinct luciferase centres with their own catalytic activities <ref name="main">PMID:15665092</ref> | ||
== Structure == | == Structure == | ||
| - | Composed of residues 868-1218, domain 3 (D3) also consists of a 20aa C-terminal unresolved domain. Containing 7 α-helices and 16 β-strands, D3 is further organized into subdomains. The main portion of the enzyme appears to be a β-barrel structure composed of 10 anti-parallel strands connected via a Gly rich sequence to a 3 helix bundle. This bundle is stabilized by a hydrophobic core region as well as a multitude of H-bonding patterns<ref name="main" />. The β-barrel structure actually has some homology with the human muscle fatty acid binding protein (m-FABP, pdb= | + | Composed of residues 868-1218, domain 3 (D3) also consists of a 20aa C-terminal unresolved domain. Containing 7 α-helices and 16 β-strands, D3 is further organized into subdomains. The main portion of the enzyme appears to be a β-barrel structure composed of 10 anti-parallel strands connected via a Gly rich sequence to a 3 helix bundle. This bundle is stabilized by a hydrophobic core region as well as a multitude of H-bonding patterns<ref name="main" />. The β-barrel structure actually has some homology with the human muscle fatty acid binding protein (m-FABP, pdb= [[1hmt]]). Both are part of a "β-clam" subdomain family, responsible for binding of hydrophobic molecules. However, other known β-clam structures do not possess enzymatic activity<ref name="main" />. |
| Line 24: | Line 21: | ||
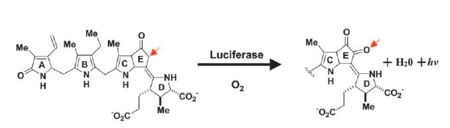
| - | [[Image:Luciferase_reaction.jpg]] | + | [[Image:Luciferase_reaction.jpg|left|thumb|450px]] |
''Image courtesy of L. Wayne Schultz.'' | ''Image courtesy of L. Wayne Schultz.'' | ||
| + | == References == | ||
| + | <references /> | ||
| + | ==Content Donors== | ||
| + | |||
| + | All the initial portions of this page were created by [[User:James Jones|James Jones]] and were moved because it deserved its own separate page distinct from the associated PDB entry. | ||
| + | </StructureSection> | ||
| + | ==3D structures of luciferase== | ||
| - | == Related Links == | ||
[[Luciferase]] | [[Luciferase]] | ||
| - | [[1vpr]] is the structure used on this page. | + | ==See Also== |
| + | *[[1vpr]] is the structure used on this page. | ||
| + | *[[Colored & Bioluminescent Proteins]] | ||
| + | * [[PyMOL]] | ||
| - | [http://www.pymol.org/ Pymol molecular viewer] | ||
| - | + | == External Resources == | |
| - | [http://www. | + | *[http://www.pdb.org/pdb/explore/explore.do?structureId=1VPR Protein Data Bank file on 1VPR] |
| - | [http:// | + | *[http://www.ncbi.nlm.nih.gov/protein/ABO61076.1?ordinalpos=1&itool=EntrezSystem2.PEntrez.Sequence.Sequence_ResultsPanel.Sequence_RVDocSum NCBI protein entry on ''P. lunula'' luciferase] |
| - | + | *[http://dx.doi.org/10.2210/rcsb_pdb/mom_2006_6 PDB molecule of the month feature on luciferases] | |
| - | + | [[Category:Topic Page]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
| |||||||||||
3D structures of luciferase
See Also
- 1vpr is the structure used on this page.
- Colored & Bioluminescent Proteins
- PyMOL
External Resources
Proteopedia Page Contributors and Editors (what is this?)
Wayne Decatur, Michal Harel, Alexander Berchansky, James Jones, Joel L. Sussman, David Canner