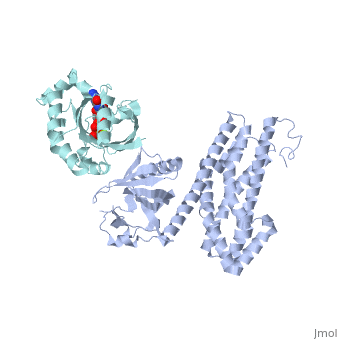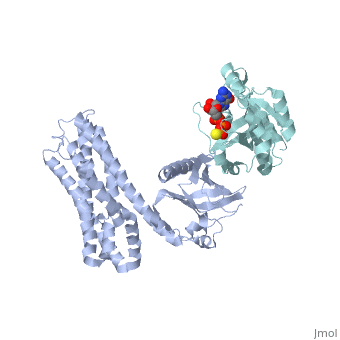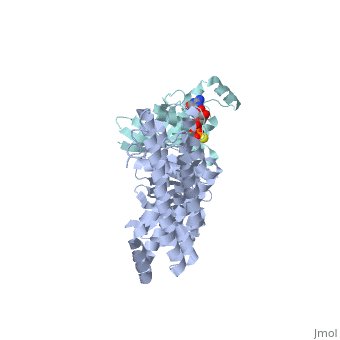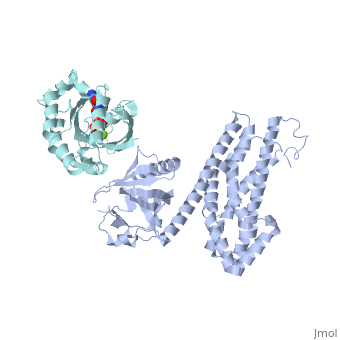|
|
| (6 intermediate revisions not shown.) |
| Line 1: |
Line 1: |
| - | {{Seed}} | |
| - | [[Image:3kz1.png|left|200px]] | |
| | | | |
| - | <!--
| + | ==Crystal Structure of the Complex of PDZ-RhoGEF DH/PH domains with GTP-gamma-S Activated RhoA== |
| - | The line below this paragraph, containing "STRUCTURE_3kz1", creates the "Structure Box" on the page.
| + | <StructureSection load='3kz1' size='340' side='right'caption='[[3kz1]], [[Resolution|resolution]] 2.70Å' scene=''> |
| - | You may change the PDB parameter (which sets the PDB file loaded into the applet)
| + | == Structural highlights == |
| - | or the SCENE parameter (which sets the initial scene displayed when the page is loaded),
| + | <table><tr><td colspan='2'>[[3kz1]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3KZ1 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=3KZ1 FirstGlance]. <br> |
| - | or leave the SCENE parameter empty for the default display.
| + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.7Å</td></tr> |
| - | --> | + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=GSP:5-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE'>GSP</scene>, <scene name='pdbligand=MG:MAGNESIUM+ION'>MG</scene></td></tr> |
| - | {{STRUCTURE_3kz1| PDB=3kz1 | SCENE= }}
| + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=3kz1 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3kz1 OCA], [https://pdbe.org/3kz1 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=3kz1 RCSB], [https://www.ebi.ac.uk/pdbsum/3kz1 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=3kz1 ProSAT]</span></td></tr> |
| | + | </table> |
| | + | == Function == |
| | + | [https://www.uniprot.org/uniprot/RHOA_HUMAN RHOA_HUMAN] Regulates a signal transduction pathway linking plasma membrane receptors to the assembly of focal adhesions and actin stress fibers. Involved in a microtubule-dependent signal that is required for the myosin contractile ring formation during cell cycle cytokinesis. Plays an essential role in cleavage furrow formation. Required for the apical junction formation of keratinocyte cell-cell adhesion. Serves as a target for the yopT cysteine peptidase from Yersinia pestis, vector of the plague, and Yersinia pseudotuberculosis, which causes gastrointestinal disorders. Stimulates PKN2 kinase activity. May be an activator of PLCE1. Activated by ARHGEF2, which promotes the exchange of GDP for GTP. Essential for the SPATA13-mediated regulation of cell migration and adhesion assembly and disassembly. The MEMO1-RHOA-DIAPH1 signaling pathway plays an important role in ERBB2-dependent stabilization of microtubules at the cell cortex. It controls the localization of APC and CLASP2 to the cell membrane, via the regulation of GSK3B activity. In turn, membrane-bound APC allows the localization of the MACF1 to the cell membrane, which is required for microtubule capture and stabilization.<ref>PMID:8910519</ref> <ref>PMID:9121475</ref> <ref>PMID:12900402</ref> <ref>PMID:16103226</ref> <ref>PMID:16236794</ref> <ref>PMID:19934221</ref> <ref>PMID:20937854</ref> <ref>PMID:20974804</ref> |
| | + | == Evolutionary Conservation == |
| | + | [[Image:Consurf_key_small.gif|200px|right]] |
| | + | Check<jmol> |
| | + | <jmolCheckbox> |
| | + | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/kz/3kz1_consurf.spt"</scriptWhenChecked> |
| | + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> |
| | + | <text>to colour the structure by Evolutionary Conservation</text> |
| | + | </jmolCheckbox> |
| | + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=3kz1 ConSurf]. |
| | + | <div style="clear:both"></div> |
| | + | <div style="background-color:#fffaf0;"> |
| | + | == Publication Abstract from PubMed == |
| | + | Guanine nucleotide exchange factors (GEFs) catalyze exchange of GDP for GTP by stabilizing the nucleotide-free state of the small GTPases through their Dbl homology/pleckstrin homology (DH.PH) domains. Unconventionally, PDZ-RhoGEF (PRG), a member of the RGS-RhoGEFs, binds tightly to both nucleotide-free and activated RhoA (RhoA.GTP). We have characterized the interaction between PRG and activated RhoA and determined the structure of the PRG-DH.PH-RhoA.GTPgammaS (guanosine 5'-O-[gamma-thio]triphosphate) complex. The interface bears striking similarity to a GTPase-effector interface and involves the switch regions in RhoA and a hydrophobic patch in PRG-PH that is conserved among all Lbc RhoGEFs. The two surfaces that bind activated and nucleotide-free RhoA on PRG-DH.PH do not overlap, and a ternary complex of PRG-DH.PH bound to both forms of RhoA can be isolated by size-exclusion chromatography. This novel interaction between activated RhoA and PH could play a key role in regulation of RhoGEF activity in vivo. |
| | | | |
| - | ===Crystal Structure of the Complex of PDZ-RhoGEF DH/PH domains with GTP-gamma-S Activated RhoA===
| + | Activated RhoA binds to the pleckstrin homology (PH) domain of PDZ-RhoGEF, a potential site for autoregulation.,Chen Z, Medina F, Liu MY, Thomas C, Sprang SR, Sternweis PC J Biol Chem. 2010 Jul 2;285(27):21070-81. Epub 2010 Apr 29. PMID:20430886<ref>PMID:20430886</ref> |
| | | | |
| | + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> |
| | + | </div> |
| | + | <div class="pdbe-citations 3kz1" style="background-color:#fffaf0;"></div> |
| | | | |
| - | <!--
| + | ==See Also== |
| - | The line below this paragraph, {{ABSTRACT_PUBMED_20430886}}, adds the Publication Abstract to the page
| + | *[[Rho GTPase 3D structures|Rho GTPase 3D structures]] |
| - | (as it appears on PubMed at http://www.pubmed.gov), where 20430886 is the PubMed ID number.
| + | *[[Rho guanine nucleotide exchange factor|Rho guanine nucleotide exchange factor]] |
| - | -->
| + | *[[Rho guanine nucleotide exchange factor 3D structures|Rho guanine nucleotide exchange factor 3D structures]] |
| - | {{ABSTRACT_PUBMED_20430886}}
| + | == References == |
| - | | + | <references/> |
| - | ==About this Structure== | + | __TOC__ |
| - | 3KZ1 is a 4 chains structure with sequences from [http://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3KZ1 OCA].
| + | </StructureSection> |
| - | | + | |
| - | ==Reference== | + | |
| - | <ref group="xtra">PMID:20430886</ref><references group="xtra"/> | + | |
| | [[Category: Homo sapiens]] | | [[Category: Homo sapiens]] |
| - | [[Category: Chen, Z.]] | + | [[Category: Large Structures]] |
| - | [[Category: Sprang, S R.]] | + | [[Category: Chen Z]] |
| - | [[Category: Sternweis, P C.]] | + | [[Category: Sprang SR]] |
| - | [[Category: Cytoskeleton]] | + | [[Category: Sternweis PC]] |
| - | [[Category: Dh]]
| + | |
| - | [[Category: Gtp-binding]]
| + | |
| - | [[Category: Gtpase activation]]
| + | |
| - | [[Category: Guanine-nucleotide releasing factor]]
| + | |
| - | [[Category: Magnesium]]
| + | |
| - | [[Category: Membrane]]
| + | |
| - | [[Category: Nucleotide-binding]]
| + | |
| - | [[Category: Ph]]
| + | |
| - | [[Category: Prenylation]]
| + | |
| - | [[Category: Proto-oncogene]]
| + | |
| - | [[Category: Regulation of rhoa gtpase]]
| + | |
| - | [[Category: Rho]]
| + | |
| - | [[Category: Rhogef]]
| + | |
| - | [[Category: Signaling protein]]
| + | |
| - | | + | |
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Wed Jul 28 12:08:04 2010''
| + | |
| Structural highlights
Function
RHOA_HUMAN Regulates a signal transduction pathway linking plasma membrane receptors to the assembly of focal adhesions and actin stress fibers. Involved in a microtubule-dependent signal that is required for the myosin contractile ring formation during cell cycle cytokinesis. Plays an essential role in cleavage furrow formation. Required for the apical junction formation of keratinocyte cell-cell adhesion. Serves as a target for the yopT cysteine peptidase from Yersinia pestis, vector of the plague, and Yersinia pseudotuberculosis, which causes gastrointestinal disorders. Stimulates PKN2 kinase activity. May be an activator of PLCE1. Activated by ARHGEF2, which promotes the exchange of GDP for GTP. Essential for the SPATA13-mediated regulation of cell migration and adhesion assembly and disassembly. The MEMO1-RHOA-DIAPH1 signaling pathway plays an important role in ERBB2-dependent stabilization of microtubules at the cell cortex. It controls the localization of APC and CLASP2 to the cell membrane, via the regulation of GSK3B activity. In turn, membrane-bound APC allows the localization of the MACF1 to the cell membrane, which is required for microtubule capture and stabilization.[1] [2] [3] [4] [5] [6] [7] [8]
Evolutionary Conservation
Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf.
Publication Abstract from PubMed
Guanine nucleotide exchange factors (GEFs) catalyze exchange of GDP for GTP by stabilizing the nucleotide-free state of the small GTPases through their Dbl homology/pleckstrin homology (DH.PH) domains. Unconventionally, PDZ-RhoGEF (PRG), a member of the RGS-RhoGEFs, binds tightly to both nucleotide-free and activated RhoA (RhoA.GTP). We have characterized the interaction between PRG and activated RhoA and determined the structure of the PRG-DH.PH-RhoA.GTPgammaS (guanosine 5'-O-[gamma-thio]triphosphate) complex. The interface bears striking similarity to a GTPase-effector interface and involves the switch regions in RhoA and a hydrophobic patch in PRG-PH that is conserved among all Lbc RhoGEFs. The two surfaces that bind activated and nucleotide-free RhoA on PRG-DH.PH do not overlap, and a ternary complex of PRG-DH.PH bound to both forms of RhoA can be isolated by size-exclusion chromatography. This novel interaction between activated RhoA and PH could play a key role in regulation of RhoGEF activity in vivo.
Activated RhoA binds to the pleckstrin homology (PH) domain of PDZ-RhoGEF, a potential site for autoregulation.,Chen Z, Medina F, Liu MY, Thomas C, Sprang SR, Sternweis PC J Biol Chem. 2010 Jul 2;285(27):21070-81. Epub 2010 Apr 29. PMID:20430886[9]
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
See Also
References
- ↑ Quilliam LA, Lambert QT, Mickelson-Young LA, Westwick JK, Sparks AB, Kay BK, Jenkins NA, Gilbert DJ, Copeland NG, Der CJ. Isolation of a NCK-associated kinase, PRK2, an SH3-binding protein and potential effector of Rho protein signaling. J Biol Chem. 1996 Nov 15;271(46):28772-6. PMID:8910519
- ↑ Vincent S, Settleman J. The PRK2 kinase is a potential effector target of both Rho and Rac GTPases and regulates actin cytoskeletal organization. Mol Cell Biol. 1997 Apr;17(4):2247-56. PMID:9121475
- ↑ Wing MR, Snyder JT, Sondek J, Harden TK. Direct activation of phospholipase C-epsilon by Rho. J Biol Chem. 2003 Oct 17;278(42):41253-8. Epub 2003 Aug 4. PMID:12900402 doi:http://dx.doi.org/10.1074/jbc.M306904200
- ↑ Yuce O, Piekny A, Glotzer M. An ECT2-centralspindlin complex regulates the localization and function of RhoA. J Cell Biol. 2005 Aug 15;170(4):571-82. PMID:16103226 doi:10.1083/jcb.200501097
- ↑ Kamijo K, Ohara N, Abe M, Uchimura T, Hosoya H, Lee JS, Miki T. Dissecting the role of Rho-mediated signaling in contractile ring formation. Mol Biol Cell. 2006 Jan;17(1):43-55. Epub 2005 Oct 19. PMID:16236794 doi:10.1091/mbc.E05-06-0569
- ↑ Bristow JM, Sellers MH, Majumdar D, Anderson B, Hu L, Webb DJ. The Rho-family GEF Asef2 activates Rac to modulate adhesion and actin dynamics and thereby regulate cell migration. J Cell Sci. 2009 Dec 15;122(Pt 24):4535-46. doi: 10.1242/jcs.053728. Epub 2009, Nov 24. PMID:19934221 doi:10.1242/jcs.053728
- ↑ Zaoui K, Benseddik K, Daou P, Salaun D, Badache A. ErbB2 receptor controls microtubule capture by recruiting ACF7 to the plasma membrane of migrating cells. Proc Natl Acad Sci U S A. 2010 Oct 26;107(43):18517-22. doi:, 10.1073/pnas.1000975107. Epub 2010 Oct 11. PMID:20937854 doi:10.1073/pnas.1000975107
- ↑ Wallace SW, Magalhaes A, Hall A. The Rho target PRK2 regulates apical junction formation in human bronchial epithelial cells. Mol Cell Biol. 2011 Jan;31(1):81-91. doi: 10.1128/MCB.01001-10. Epub 2010 Oct 25. PMID:20974804 doi:10.1128/MCB.01001-10
- ↑ Chen Z, Medina F, Liu MY, Thomas C, Sprang SR, Sternweis PC. Activated RhoA binds to the pleckstrin homology (PH) domain of PDZ-RhoGEF, a potential site for autoregulation. J Biol Chem. 2010 Jul 2;285(27):21070-81. Epub 2010 Apr 29. PMID:20430886 doi:10.1074/jbc.M110.122549
|