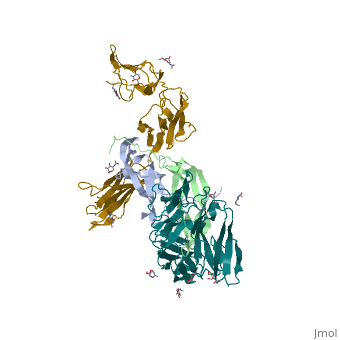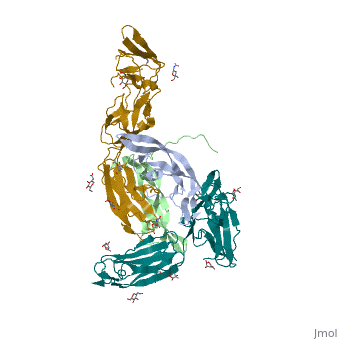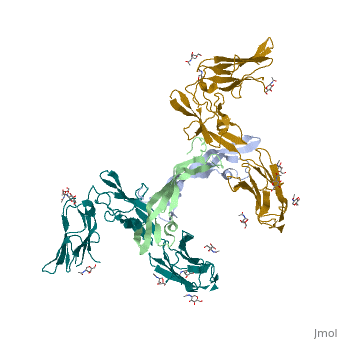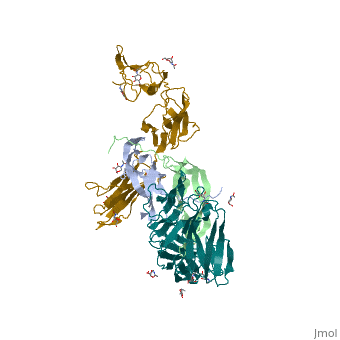
We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
3mjg
From Proteopedia
(Difference between revisions)
| (8 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | {{Seed}} | ||
| - | [[Image:3mjg.png|left|200px]] | ||
| - | < | + | ==The structure of a platelet derived growth factor receptor complex== |
| - | + | <StructureSection load='3mjg' size='340' side='right'caption='[[3mjg]], [[Resolution|resolution]] 2.30Å' scene=''> | |
| - | You may | + | == Structural highlights == |
| - | + | <table><tr><td colspan='2'>[[3mjg]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3MJG OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=3MJG FirstGlance]. <br> | |
| - | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.3Å</td></tr> | |
| - | -- | + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=NAG:N-ACETYL-D-GLUCOSAMINE'>NAG</scene>, <scene name='pdbligand=NDG:2-(ACETYLAMINO)-2-DEOXY-A-D-GLUCOPYRANOSE'>NDG</scene></td></tr> |
| - | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=3mjg FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3mjg OCA], [https://pdbe.org/3mjg PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=3mjg RCSB], [https://www.ebi.ac.uk/pdbsum/3mjg PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=3mjg ProSAT]</span></td></tr> | |
| + | </table> | ||
| + | == Disease == | ||
| + | [https://www.uniprot.org/uniprot/PDGFB_HUMAN PDGFB_HUMAN] Note=A chromosomal aberration involving PDGFB is found in dermatofibrosarcoma protuberans. Translocation t(17;22)(q22;q13) with PDGFB.<ref>PMID:12660034</ref> | ||
| + | == Function == | ||
| + | [https://www.uniprot.org/uniprot/PDGFB_HUMAN PDGFB_HUMAN] Growth factor that plays an essential role in the regulation of embryonic development, cell proliferation, cell migration, survival and chemotaxis. Potent mitogen for cells of mesenchymal origin. Required for normal proliferation and recruitment of pericytes and vascular smooth muscle cells in the central nervous system, skin, lung, heart and placenta. Required for normal blood vessel development, and for normal development of kidney glomeruli. Plays an important role in wound healing. Signaling is modulated by the formation of heterodimers with PDGFA (By similarity). | ||
| + | == Evolutionary Conservation == | ||
| + | [[Image:Consurf_key_small.gif|200px|right]] | ||
| + | Check<jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/mj/3mjg_consurf.spt"</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview03.spt</scriptWhenUnchecked> | ||
| + | <text>to colour the structure by Evolutionary Conservation</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=3mjg ConSurf]. | ||
| + | <div style="clear:both"></div> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
| + | Platelet-derived growth factors (PDGFs) and their receptors (PDGFRs) are prototypic growth factors and receptor tyrosine kinases which have critical functions in development. We show that PDGFs share a conserved region in their prodomain sequences which can remain noncovalently associated with the mature cystine-knot growth factor domain after processing. The structure of the PDGF-A/propeptide complex reveals this conserved, hydrophobic association mode. We also present the structure of the complex between PDGF-B and the first three Ig domains of PDGFRbeta, showing that two PDGF-B protomers clamp PDGFRbeta at their dimerization seam. The PDGF-B:PDGFRbeta interface is predominantly hydrophobic, and PDGFRs and the PDGF propeptides occupy overlapping positions on mature PDGFs, rationalizing the need of propeptides by PDGFs to cover functionally important hydrophobic surfaces during secretion. A large-scale structural organization and rearrangement is observed for PDGF-B upon receptor binding, in which the PDGF-B L1 loop, disordered in the structure of the free form, adopts a highly specific conformation to form hydrophobic interactions with the third Ig domain of PDGFRbeta. Calorimetric data also shows that the membrane-proximal homotypic PDGFRalpha interaction, albeit required for activation, contributes negatively to ligand binding. The structural and biochemical data together offer insights into PDGF-PDGFR signaling, as well as strategies for PDGF-antagonism. | ||
| - | + | Structures of a platelet-derived growth factor/propeptide complex and a platelet-derived growth factor/receptor complex.,Shim AH, Liu H, Focia PJ, Chen X, Lin PC, He X Proc Natl Acad Sci U S A. 2010 Jun 22;107(25):11307-12. Epub 2010 Jun 2. PMID:20534510<ref>PMID:20534510</ref> | |
| - | + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |
| - | + | </div> | |
| - | + | <div class="pdbe-citations 3mjg" style="background-color:#fffaf0;"></div> | |
| - | + | == References == | |
| - | + | <references/> | |
| - | + | __TOC__ | |
| - | + | </StructureSection> | |
| - | == | + | |
| - | + | ||
| - | + | ||
| - | == | + | |
| - | < | + | |
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
| - | [[Category: | + | [[Category: Large Structures]] |
| - | [[Category: He | + | [[Category: He X]] |
| - | [[Category: Shim | + | [[Category: Shim AHR]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
The structure of a platelet derived growth factor receptor complex
| |||||||||||