T4 RNA ligase 2 (Rnl2)
From Proteopedia
(Difference between revisions)
| (3 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='1s68' size='450' side='right' scene='Sandbox_157/T4_rnl2_active_site/12' caption=''> | |
| + | |||
== Overview: == | == Overview: == | ||
| Line 14: | Line 15: | ||
== Structural Components Contributing to the Active Site: == | == Structural Components Contributing to the Active Site: == | ||
| + | <scene name='Sandbox_157/T4_rnl2_active_site/13'>Structural components contributing to the Rnl2 active site</scene> | ||
| - | <applet load='1s68' size='350' frame='true' align='left' caption='Structural components contributing to the Rnl2 active site' scene='Sandbox_157/T4_rnl2_active_site/13'/> | ||
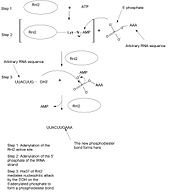
Above the components of the actual active site were discussed in relation to the function of the protein, however there are several indirect structural components which are linked to maintaining active site structure as well as aiding in conformational changes while it carries out its function. For instance <scene name='Sandbox_157/T4_rnl2_active_site/26'>Asp 120 (silver)</scene> of motif IIIa (pink), located next to Phe 119 (see above for function), serves as a focal point of hydrophilic contacts which include a bidentate salt bridge to <scene name='Sandbox_157/T4_rnl2_active_site/27'>Arg 221 (dark green)</scene> and a hydrogen bond to <scene name='Sandbox_157/T4_rnl2_active_site/28'> Tyr 5 (dark purple)</scene>. These <scene name='Sandbox_157/T4_rnl2_active_site/22'>structural components tether 4 beta strands that make up the nucleotide binding pocket of Rnl2. </scene> <scene name='Sandbox_157/T4_rnl2_active_site/23'>click here</scene> to see H-bonding patterns between side chains <ref name="lu"/>. <scene name='Sandbox_157/T4_rnl2_active_site/29'>Glu 34 (dark red)</scene> has also been found to be structurally important, although not making direct contact with AMP, as it functions to make sure that the loop containing the lysine nucleophile is in proper conformation for adenylation <ref name="lu"/>. It accomplishes this structural function by tethering the backside of the motif I loop (green) to the start of<scene name='Sandbox_157/T4_rnl2_active_site/24'>alpha helix 6 (lime green)</scene> through a hydrogen bond to Ser 170 (dark purple) <ref name="lu"/>. | Above the components of the actual active site were discussed in relation to the function of the protein, however there are several indirect structural components which are linked to maintaining active site structure as well as aiding in conformational changes while it carries out its function. For instance <scene name='Sandbox_157/T4_rnl2_active_site/26'>Asp 120 (silver)</scene> of motif IIIa (pink), located next to Phe 119 (see above for function), serves as a focal point of hydrophilic contacts which include a bidentate salt bridge to <scene name='Sandbox_157/T4_rnl2_active_site/27'>Arg 221 (dark green)</scene> and a hydrogen bond to <scene name='Sandbox_157/T4_rnl2_active_site/28'> Tyr 5 (dark purple)</scene>. These <scene name='Sandbox_157/T4_rnl2_active_site/22'>structural components tether 4 beta strands that make up the nucleotide binding pocket of Rnl2. </scene> <scene name='Sandbox_157/T4_rnl2_active_site/23'>click here</scene> to see H-bonding patterns between side chains <ref name="lu"/>. <scene name='Sandbox_157/T4_rnl2_active_site/29'>Glu 34 (dark red)</scene> has also been found to be structurally important, although not making direct contact with AMP, as it functions to make sure that the loop containing the lysine nucleophile is in proper conformation for adenylation <ref name="lu"/>. It accomplishes this structural function by tethering the backside of the motif I loop (green) to the start of<scene name='Sandbox_157/T4_rnl2_active_site/24'>alpha helix 6 (lime green)</scene> through a hydrogen bond to Ser 170 (dark purple) <ref name="lu"/>. | ||
| Line 24: | Line 25: | ||
Trypanosomaisis affects between 300, 000 to 500, 000 people in rural parts of Central Africa every year in which almost all cases are fatal and currently no effective treatment method exists for the disease.<ref>PMID:16020726 | Trypanosomaisis affects between 300, 000 to 500, 000 people in rural parts of Central Africa every year in which almost all cases are fatal and currently no effective treatment method exists for the disease.<ref>PMID:16020726 | ||
</ref> Because of the several similarities Rnl2 shares with REL-1 and other RELs, in particular its active site and binding sites, an understanding of this protein's structure in relation to its function can give further insight as to how the REL's structure may relate to their function. If the enzymes behave similar mechanistically, target pharmaceuticals designed to inhibit the Rnl2 active site or binding sites may also serve as potential inhibitors against RELs. Since the RELs have been found to be essential to trypanosomiasis and leishmaniasis, mentioned earlier, they have become prime targets for drugs against these microorganisms <ref name="lu"/>. Such drugs can be designed to mimic the substrate, bind to impair the AMP binding pocket, or bind to other surface residues which are essential to the function of the protein such as Glu 34, or His 37 <ref name="lu"/>. | </ref> Because of the several similarities Rnl2 shares with REL-1 and other RELs, in particular its active site and binding sites, an understanding of this protein's structure in relation to its function can give further insight as to how the REL's structure may relate to their function. If the enzymes behave similar mechanistically, target pharmaceuticals designed to inhibit the Rnl2 active site or binding sites may also serve as potential inhibitors against RELs. Since the RELs have been found to be essential to trypanosomiasis and leishmaniasis, mentioned earlier, they have become prime targets for drugs against these microorganisms <ref name="lu"/>. Such drugs can be designed to mimic the substrate, bind to impair the AMP binding pocket, or bind to other surface residues which are essential to the function of the protein such as Glu 34, or His 37 <ref name="lu"/>. | ||
| + | </StructureSection> | ||
| + | __NOTOC__ | ||
| + | ==3D structures of RNA ligase== | ||
| - | + | [[RNA ligase]] | |
Current revision
| |||||||||||
3D structures of RNA ligase
References:
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 1.19 1.20 1.21 1.22 1.23 1.24 Ho CK, Wang LK, Lima CD, Shuman S. Structure and mechanism of RNA ligase. Structure. 2004 Feb;12(2):327-39. PMID:14962393 doi:10.1016/j.str.2004.01.011
- ↑ Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, Bohme U, Hannick L, Aslett MA, Shallom J, Marcello L, Hou L, Wickstead B, Alsmark UC, Arrowsmith C, Atkin RJ, Barron AJ, Bringaud F, Brooks K, Carrington M, Cherevach I, Chillingworth TJ, Churcher C, Clark LN, Corton CH, Cronin A, Davies RM, Doggett J, Djikeng A, Feldblyum T, Field MC, Fraser A, Goodhead I, Hance Z, Harper D, Harris BR, Hauser H, Hostetler J, Ivens A, Jagels K, Johnson D, Johnson J, Jones K, Kerhornou AX, Koo H, Larke N, Landfear S, Larkin C, Leech V, Line A, Lord A, Macleod A, Mooney PJ, Moule S, Martin DM, Morgan GW, Mungall K, Norbertczak H, Ormond D, Pai G, Peacock CS, Peterson J, Quail MA, Rabbinowitsch E, Rajandream MA, Reitter C, Salzberg SL, Sanders M, Schobel S, Sharp S, Simmonds M, Simpson AJ, Tallon L, Turner CM, Tait A, Tivey AR, Van Aken S, Walker D, Wanless D, Wang S, White B, White O, Whitehead S, Woodward J, Wortman J, Adams MD, Embley TM, Gull K, Ullu E, Barry JD, Fairlamb AH, Opperdoes F, Barrell BG, Donelson JE, Hall N, Fraser CM, Melville SE, El-Sayed NM. The genome of the African trypanosome Trypanosoma brucei. Science. 2005 Jul 15;309(5733):416-22. PMID:16020726 doi:309/5733/416
Proteopedia Page Contributors and Editors (what is this?)
William Eisbrenner, Alexander Berchansky, David Canner, Sonja Senekovic, Michal Harel, Andrea Gorrell