Ubiquitin Structure & Function
From Proteopedia
(Difference between revisions)
| (10 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | [[Ubiquitin]] is a single 8565 M<sub>r</sub> polypeptide consisting of 76 amino acid residues | + | <StructureSection load='1ubq' size='350' side='right' caption='Human ubiquitin, [[1ubq]])' scene=''> |
| - | + | ||
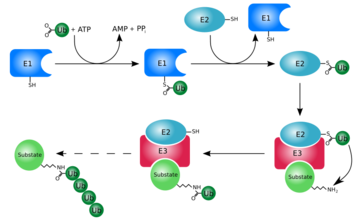
| + | [[Ubiquitin]] is a single 8565 M<sub>r</sub> polypeptide consisting of 76 amino acid residues and is highly known for its role in ATP-dependent protein degradation<ref name="mainpaper">PMID: 3041007</ref> | ||
| + | |||
=Introduction= | =Introduction= | ||
| - | Ubiquitin is one of the most highly conserved eukaryotic proteins. Primary structures found throughout ubiquitin are identical in all bovine, insects and human ubiquitin <ref name="mainpaper"/>. The only difference observed amongst these species is seen in the terminal Gly-Gly residues. Yeast and oat ubiquitin only differ in three of the 76 residues when compared to ubiquitin found in higher eukaryotes<ref name="mainpaper"/>.[[image:1ubiq.png| thumb |none | upright=2.0 |Ubiquitin structure: Arg74 in pink and Gly75 Gly76 in white.]] | + | Ubiquitin is one of the most highly conserved eukaryotic proteins. Primary structures found throughout ubiquitin are identical in all bovine, insects and human ubiquitin <ref name="mainpaper"/>. The only difference observed amongst these species is seen in the terminal Gly-Gly residues. Yeast and oat ubiquitin only differ in three of the 76 residues when compared to ubiquitin found in higher eukaryotes<ref name="mainpaper"/>. |
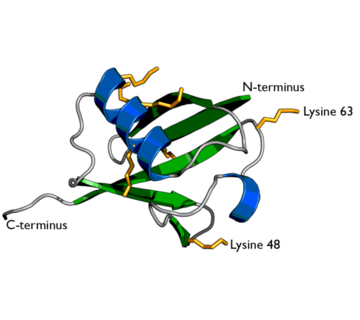
| + | [[image:1ubiq.png| thumb |none | upright=2.0 |Ubiquitin structure: Arg74 in pink and Gly75 Gly76 in white.]] | ||
| + | |||
| + | <!-- The content below was inserted by the ConSurf template --> | ||
| + | == Evolutionary Conservation == | ||
| + | [[Image:Consurf_key_small.gif|right|200px]] | ||
| + | Check | ||
| + | <jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked> | ||
| + | select protein; define ~consurf_to_do selected; | ||
| + | consurf_initial_scene = true; | ||
| + | script /wiki/ConSurf/ub/1ubq_consurf.spt; | ||
| + | </scriptWhenChecked> | ||
| + | <scriptWhenUnchecked> | ||
| + | script /wiki/extensions/Proteopedia/spt/initialview01.spt; | ||
| + | </scriptWhenUnchecked> | ||
| + | <text>to colour the structure by Evolutionary Conservation</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. | ||
| + | You may read the [[Conservation%2C_Evolutionary|explanation]] | ||
| + | of the method and the full data available from | ||
| + | [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1ubq ConSurf]. | ||
| + | <!-- end of content inserted by the ConSurf template --> | ||
| + | |||
Ubiquitin can not only be found in the nucleus, but in the cytoplasm and cell-surface membrane as well<ref name="mainpaper"/>. | Ubiquitin can not only be found in the nucleus, but in the cytoplasm and cell-surface membrane as well<ref name="mainpaper"/>. | ||
| Line 34: | Line 60: | ||
There are numerous diseases that may develop as a result of ubiquitin abnormalities. There are two disease categories possible in non-lethal states. One being the result of function loss and the other being function gain. Loss of function may occur due to a target substrate mutation or a mutation in a ubiquitin enzyme causing protein stabilization and a decrease in protein degradation. Function gain, on the other hand, results in an increase in protein degradation. | There are numerous diseases that may develop as a result of ubiquitin abnormalities. There are two disease categories possible in non-lethal states. One being the result of function loss and the other being function gain. Loss of function may occur due to a target substrate mutation or a mutation in a ubiquitin enzyme causing protein stabilization and a decrease in protein degradation. Function gain, on the other hand, results in an increase in protein degradation. | ||
Cancer may result from either case. Oncoproteins may become stabilized while tumor suppressor genes may become destabilized. Liddle's Syndrome is a type of early-onset hypertension<ref name="liddles">PMID: 8521520</ref>. Sodium ions and water are excessively reabsorbed caused by E3 ligase non-recognition. Angleman syndrome is caused by a E3 ligase defect. This defect causes affects in human brain development resulting in symptoms such as mental retardation, seizures and abnormal gait. Lastly, neurogenetive diseases are caused by the accumulation of ubiquitin-conjugates. Diseases of this nature include Alzheimers and Parkinson's. | Cancer may result from either case. Oncoproteins may become stabilized while tumor suppressor genes may become destabilized. Liddle's Syndrome is a type of early-onset hypertension<ref name="liddles">PMID: 8521520</ref>. Sodium ions and water are excessively reabsorbed caused by E3 ligase non-recognition. Angleman syndrome is caused by a E3 ligase defect. This defect causes affects in human brain development resulting in symptoms such as mental retardation, seizures and abnormal gait. Lastly, neurogenetive diseases are caused by the accumulation of ubiquitin-conjugates. Diseases of this nature include Alzheimers and Parkinson's. | ||
| + | |||
| + | Infectious agents can manipulate ubiquitin or deubiquitination and one such protein is Chlamydia trachomatis. Chlamydia trachomatis' protein Cdu-1 catalyzes the hydrolysis of ubiquitin chains from Mcl-1. When polyubiquitnated, Mcl-1 is destined to be degraded by the proteasome, lowering the level of Mcl-1 and subsequently leading to apoptosis. The activity of Cdu-1 counteracts this by removing the ubiquitin, thus leading to higher levels of Mcl-1 in the cell. Additional information can be found here [[User:Karsten Theis/5B5Q]] | ||
| + | |||
| + | SEE ALSO [[Tumor susceptibility gene 101]] | ||
| + | </StructureSection> | ||
| + | |||
| + | ==3D structures of ubiqitin== | ||
| + | |||
| + | [[Ubiquitin]] | ||
| + | |||
=References= | =References= | ||
<references/> | <references/> | ||
Current revision
| |||||||||||
3D structures of ubiqitin
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 A resolution. J Mol Biol. 1987 Apr 5;194(3):531-44. PMID:3041007
- ↑ 2.0 2.1 Vijay-Kumar S, Bugg CE, Wilkinson KD, Cook WJ. Three-dimensional structure of ubiquitin at 2.8 A resolution. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3582-5. PMID:2987935
- ↑ Cox MJ, Haas AL, Wilkinson KD. Role of ubiquitin conformations in the specificity of protein degradation: iodinated derivatives with altered conformations and activities. Arch Biochem Biophys. 1986 Nov 1;250(2):400-9. PMID:3022650
- ↑ Wilkinson KD. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997 Dec;11(14):1245-56. PMID:9409543
- ↑ Hochstrasser, M. 1996. Ubiquitin-dependent protein Degradation. Annu Rev Genet. 30: 405-439
- ↑ Snyder PM, Price MP, McDonald FJ, Adams CM, Volk KA, Zeiher BG, Stokes JB, Welsh MJ. Mechanism by which Liddle's syndrome mutations increase activity of a human epithelial Na+ channel. Cell. 1995 Dec 15;83(6):969-78. PMID:8521520
Proteopedia Page Contributors and Editors (what is this?)
Jaclyn Gordon, Joel L. Sussman, Michal Harel, David Canner, Andrea Gorrell, Alexander Berchansky, Karsten Theis