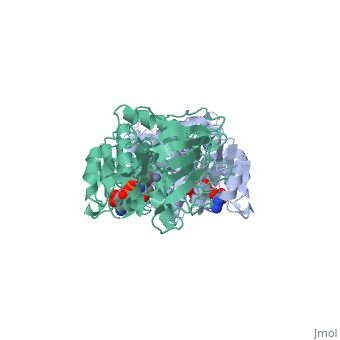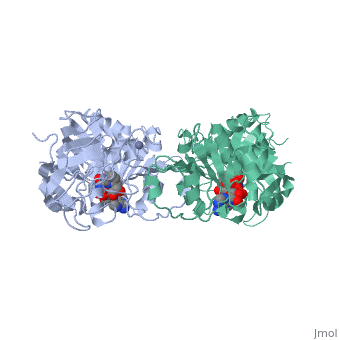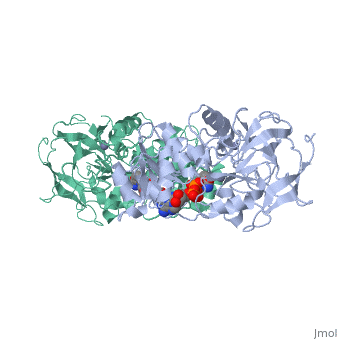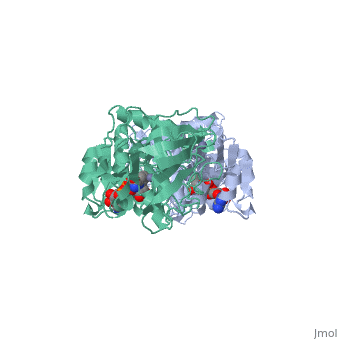
Horse Liver Alcohol Dehydrogenase
From Proteopedia
(Difference between revisions)
(→References) |
|||
| (3 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='1a72' size='450' side='right' scene='' caption='Horse liver alcohol dehydrogenase complex with Zn+2 ion (grey) and ribofuranosylpicolinamide adenine dinucleotide (PDB code [[1a72]])'> | |
===''' General Information'''=== | ===''' General Information'''=== | ||
---- | ---- | ||
| - | Horse liver alcohol dehydrogenase (LADH) is found in the liver of the species Equus Caballus (horse). It is a dimeric zinc-dependant protein, with a broad substrate specificity, being active on a variety of primary, secondary, branched, and cyclic alcohols.<ref name="structure">PMID: 8172897 </ref> The enzymatic mechanism and structures of various complexes have been extensively studied.<ref name=" tunneling"/><ref name=" active"/><ref name=" unmasking"/> The catalytic reaction is ordered, with NAD+ or NADH binding to free enzyme when primary alcohols and aldehydes, such as ethanol and acetaldehyde or benzyl alcohol and benzaldehyde, are the substrates.<ref name="structure"/>The enzyme has a molecular weight of 80 000 and is a dimmer of two identical subunits, each of which has a coenzyme binding and a catalytic domain. The coenzyme-binding domains have structures similar to those found in several other NAD-dependent dehydrogenases.<ref name="structure"/> | + | '''Horse liver alcohol dehydrogenase''' (LADH) is found in the liver of the species Equus Caballus (horse). It is a dimeric zinc-dependant protein, with a broad substrate specificity, being active on a variety of primary, secondary, branched, and cyclic alcohols.<ref name="structure">PMID: 8172897 </ref> The enzymatic mechanism and structures of various complexes have been extensively studied.<ref name=" tunneling"/><ref name=" active"/><ref name=" unmasking"/> The catalytic reaction is ordered, with NAD+ or NADH binding to free enzyme when primary alcohols and aldehydes, such as ethanol and acetaldehyde or benzyl alcohol and benzaldehyde, are the substrates.<ref name="structure"/>The enzyme has a molecular weight of 80 000 and is a dimmer of two identical subunits, each of which has a coenzyme binding and a catalytic domain. The coenzyme-binding domains have structures similar to those found in several other NAD-dependent dehydrogenases.<ref name="structure"/> |
===''' Protein Structure '''=== | ===''' Protein Structure '''=== | ||
| Line 13: | Line 13: | ||
The catalytic domain is mainly built up from three distinct antiparallel pleated-sheet regions. Residues within this domain provide ligands to the catalytic <scene name='Sandbox_163/Zinc/2' target='zinc'>zinc</scene> atom; Cys46, His67 and Cys174.<ref name=" zinc">PMID: 19177216</ref> An approximate tetrahedral coordination of this zinc is completed by a water molecule or hydroxyl ion depending on the pH. Residues 95 to 113 form a lobe that binds the second zinc atom of the subunit. This zinc is liganded in a distorted tetrahedral arrangement by four sulphur atoms from the cysteine residues 97, 100, 103 and 111. The lobe forms one side of a significant cleft in the enzyme surface suggesting that this region might constitute a second catalytic centre of unknown function.<ref name=" zinc"/> | The catalytic domain is mainly built up from three distinct antiparallel pleated-sheet regions. Residues within this domain provide ligands to the catalytic <scene name='Sandbox_163/Zinc/2' target='zinc'>zinc</scene> atom; Cys46, His67 and Cys174.<ref name=" zinc">PMID: 19177216</ref> An approximate tetrahedral coordination of this zinc is completed by a water molecule or hydroxyl ion depending on the pH. Residues 95 to 113 form a lobe that binds the second zinc atom of the subunit. This zinc is liganded in a distorted tetrahedral arrangement by four sulphur atoms from the cysteine residues 97, 100, 103 and 111. The lobe forms one side of a significant cleft in the enzyme surface suggesting that this region might constitute a second catalytic centre of unknown function.<ref name=" zinc"/> | ||
| - | < | + | |
| + | <scene name='Sandbox_163/Alcohol_dehydrogenase_ladh/6'>Highlight of Secondary Strucutres within Horse Liver Alcohol Dehydrogenase</scene> | ||
The two domains of the subunit are separated by a crevice that contains a wide and deep hydrophobic pocket. The catalytic zinc atom is at the bottom of this pocket, with the zinc-bound water molecule projecting out into the pocket. This water molecule is hydrogen-bonded to the side chain of Ser48 which in turn is hydrogen-bonded to His51.<ref name=" unmasking"/> The pocket which in all probability is the binding site for the substrate and the nicotinamide moiety of the coenzyme, is lined almost exclusively with hydrophobic side chains.<ref name=" active">PMID: 9649310 </ref><ref name=" unmasking"/> The only accessible polar groups in the vicinity of the catalytic centre are Ser48 and Thr178 apart from zinc and the zinc-bound water molecule.<ref name=" zinc"/> | The two domains of the subunit are separated by a crevice that contains a wide and deep hydrophobic pocket. The catalytic zinc atom is at the bottom of this pocket, with the zinc-bound water molecule projecting out into the pocket. This water molecule is hydrogen-bonded to the side chain of Ser48 which in turn is hydrogen-bonded to His51.<ref name=" unmasking"/> The pocket which in all probability is the binding site for the substrate and the nicotinamide moiety of the coenzyme, is lined almost exclusively with hydrophobic side chains.<ref name=" active">PMID: 9649310 </ref><ref name=" unmasking"/> The only accessible polar groups in the vicinity of the catalytic centre are Ser48 and Thr178 apart from zinc and the zinc-bound water molecule.<ref name=" zinc"/> | ||
| Line 34: | Line 35: | ||
====Additional Functions==== | ====Additional Functions==== | ||
While LADH is generally associated with the interconversion of alcohols and aldehydes, it has also received some attention for its catalysis of aldehyde. Calculations suggest that it is possible that LADH could catalyze hydration of an aldehyde when hydroxide is ligated to the active site zinc. Molecular dynamics results suggest that the geometry is favorable for nucleophilic attack of zinc bound hydroxide ion on the aldehyde carbonyl carbon.<ref name=" aldehyde">PMID: 8703951 </ref> | While LADH is generally associated with the interconversion of alcohols and aldehydes, it has also received some attention for its catalysis of aldehyde. Calculations suggest that it is possible that LADH could catalyze hydration of an aldehyde when hydroxide is ligated to the active site zinc. Molecular dynamics results suggest that the geometry is favorable for nucleophilic attack of zinc bound hydroxide ion on the aldehyde carbonyl carbon.<ref name=" aldehyde">PMID: 8703951 </ref> | ||
| + | </StructureSection> | ||
| + | __NOTOC__ | ||
| + | |||
| + | ===3D structures of horse liver alcohol dehydrogenase=== | ||
| + | [[Alcohol dehydrogenase]] | ||
== References == | == References == | ||
<references/> | <references/> | ||
Current revision
| |||||||||||
3D structures of horse liver alcohol dehydrogenase
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Ramaswamy S, Eklund H, Plapp BV. Structures of horse liver alcohol dehydrogenase complexed with NAD+ and substituted benzyl alcohols. Biochemistry. 1994 May 3;33(17):5230-7. PMID:8172897
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Bahnson BJ, Colby TD, Chin JK, Goldstein BM, Klinman JP. A link between protein structure and enzyme catalyzed hydrogen tunneling. Proc Natl Acad Sci U S A. 1997 Nov 25;94(24):12797-802. PMID:9371755
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 Colby TD, Bahnson BJ, Chin JK, Klinman JP, Goldstein BM. Active site modifications in a double mutant of liver alcohol dehydrogenase: structural studies of two enzyme-ligand complexes. Biochemistry. 1998 Jun 30;37(26):9295-304. PMID:9649310 doi:10.1021/bi973184b
- ↑ 4.0 4.1 4.2 4.3 4.4 Bahnson BJ, Park DH, Kim K, Plapp BV, Klinman JP. Unmasking of hydrogen tunneling in the horse liver alcohol dehydrogenase reaction by site-directed mutagenesis. Biochemistry. 1993 Jun 1;32(21):5503-7. PMID:8504071
- ↑ 5.0 5.1 5.2 Eklund H, Nordstrom B, Zeppezauer E, Soderlund G, Ohlsson I, Boiwe T, Soderberg BO, Tapia O, Branden CI, Akeson A. Three-dimensional structure of horse liver alcohol dehydrogenase at 2-4 A resolution. J Mol Biol. 1976 Mar 25;102(1):27-59. PMID:178875
- ↑ 6.0 6.1 6.2 Brandt EG, Hellgren M, Brinck T, Bergman T, Edholm O. Molecular dynamics study of zinc binding to cysteines in a peptide mimic of the alcohol dehydrogenase structural zinc site. Phys Chem Chem Phys. 2009 Feb 14;11(6):975-83. Epub 2008 Dec 12. PMID:19177216 doi:10.1039/b815482a
- ↑ Olson LP, Luo J, Almarsson O, Bruice TC. Mechanism of aldehyde oxidation catalyzed by horse liver alcohol dehydrogenase. Biochemistry. 1996 Jul 30;35(30):9782-91. PMID:8703951 doi:10.1021/bi952020x
Proteopedia Page Contributors and Editors (what is this?)
Meghan Hatcher, Alexander Berchansky, David Canner, Michal Harel, Simmi Parhar, Andrea Gorrell, Eran Hodis