Sandbox 30
From Proteopedia
| (38 intermediate revisions not shown.) | |||
| Line 2: | Line 2: | ||
{{Template:Oberholser_Sandbox_Reservation}} | {{Template:Oberholser_Sandbox_Reservation}} | ||
<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | <!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
| - | + | = '''Papain''' = | |
| - | = ''' | + | Note: For some reason, using the "hydrogen bonds" link in the structure section breaks the jmol display and results in scenes after that point having random disulfide bonds and solvent molecules floating in mid-air. I've tried to fix it but to no avail, so if this happens just refresh the page and everything should work again. Oh look, now the new h-bond link in the substrate binding section does the same thing. If you see disembodied floating solvent molecules after clicking it, just refresh the page. |
| - | + | ||
| - | + | ||
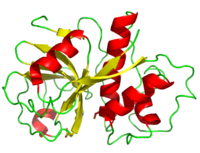
==Overview== | ==Overview== | ||
| - | + | [[Image:Papain_cartoon.png|200px|left|thumb|]] | |
| - | + | <applet load='9PAP' size='400' frame='true' align='right' scene='Sandbox_30/Papain_default/7' caption='Click on the links to the left to view different structural aspects. PDB code for this 1.65 Å resolution structure is 9PAP.' /> | |
| + | Papain is a 23.4 kDa, 212 residue cysteine endopeptidase originating from the fruit of ''Carica papaya'', where it is present in significant amounts along with three other cysteine proteases, chymopapain, glycyl endopeptidase, and caricain<ref name="9PAP PDB">[http://www.pdb.org/pdb/explore/explore.do?structureId=9PAP] 9PAP PDB</ref><ref name="sigma">[http://www.sigmaaldrich.com/life-science/metabolomics/enzyme-explorer/analytical-enzymes/papain.html] Sigma Aldrich</ref><ref name="worthington">[http://www.worthington-biochem.com/pap/default.html] Worthington Biochemical Corporation</ref>. Its action was first described by G.C. Roy in 1873. It was studied intensively from the 1950s to the 1960s, during which time it became the second enzyme ever to have its structure determined by x-ray crystallography. Finally, high resolution structural analysis in the 1980s allowed an accurate description of the enzyme's active site<ref name="worthington" />. Papain displays very wide hydrolase activity, serving as a general amidase and esterase in addition to its protease activity. As a protease, papain can hydrolyze bonds of basic amino acids, leucine, and glycine. It shows preference for residues preceded by a large hydrophobic residue, but will not cleave if valine is present on the carboxyl side of a potential cleavage site. In addition to being a very non-specific enzyme, papain is also unusually heat resistant, with maximal activity occurring at a temperature of 65° C. These properties have led to use of papain in a large variety areas. One of these areas is biological research, where papain is utilized in cell isolation. It is also useful in immunological techniques because of its ability to cleave the connection between the crystallizable fragment domain and the immunoglobulin domain of antibodies<ref name="sigma" />. Additionally, papain has found use outside of research as an inflammation control agent, a digestive aid, and even a meat tenderizer<ref>[http://www.webmd.com/vitamins-supplements/ingredientmono-69-PAPAIN.aspx?activeIngredientId=69&activeIngredientName=PAPAIN] WebMD</ref>. | ||
| - | ==Structure== | ||
| - | <applet load='1QLQ' size='450' frame='true' align='right' caption='Click on the links to the left to view different structural aspects. Ligand shown: SO4' /> | ||
| - | Trypsin's primary amino acid sequence (RPDFCLEPPYAGACRARIIRYFYNAKAGLCQTFVYGGCRAKRNNFKSAEDCLRTCGGA) <ref>[http://bip.weizmann.ac.il/oca-bin/send-seq?1qlq_A 1qlq]</ref> forms the <scene name='Sandbox_30/Backbone/1'>backbone</scene> of the protein, which then folds into secondary structures, consisting of two <scene name='Sandbox_30/Helixs_maroon/3'>α helices</scene> and two <scene name='Sandbox_30/Sheets_green/4'>β sheets</scene>. Both of the α helices are right handed and the β sheets are anti-parallel. The order of the secondary structures is easily visible when using the <scene name='Sandbox_30/Trypsin_cartoon_rainbow/2'>rainbow coloration</scene> scheme to identify secondary structures. The N-terminus (blue) is the beginning of trypsin and the C-terminus (agua-green) is the end. | ||
| - | ==Polar and Nonpolar Residues== | ||
| - | Polar residues are typically hydrophobic, and seek to be sheltered from the aqueous environments that proteins typically inhibit. The polarity of an amino acid is determined by its <scene name='Sandbox_30/Side_chains/1'>side chain</scene> (orange). When considering the <scene name='Sandbox_30/Polar_and_nonpolar/1'>ball and stick model</scene> it may look like the polar (blue) and nonpolar (crimson) residues are not organized in a specific manner, but when you consider the <scene name='Sandbox_30/Polar_and_nonpolar/2'>space filling model,</scene> it is evident that the majority of the nonpolar residues are shielded by the polar residues. | ||
| - | Another way to show this principle is by looking at the location of the <scene name='Sandbox_30/Hydrophobic_red/1'>hydrophobic sections</scene> of Trypsin (red). Alternatively, for a more in depth analysis of trypsin, you can view | ||
| - | <scene name='Sandbox_30/Space_fill_charge_and_polar/1'>charged (Blue +)(Red -), uncharged polar (purple), and hydrophobic (gray) space filling rendering</scene> which can be even more informing. The hydrophobic portions desire to be shielded from the water in the smallest area possible in order to minimize its interaction with water, thereby maximizing the entropy of the water. It is evident that basically all water molecules are kept outside the protein when viewing a <scene name='Sandbox_30/Ball_and_stick_with_water/1'>rendering with water</scene> (water-blue, trypsin-orange). This form of trypsin (PDB 1QLQ), has been modified to help enable its crystalization, and thus has four water molecules inside of it instead of the normal three which is present in the wild-type trpsin<ref> Czapinska, Honorata et al. "High-resolution structure of bovine pancreatic trypsin inhibitor with altered binding loop sequence." ''Journal of Molecular Biology.'' Volume 295, Issue 5, 4 February 2000, Pages 1237-1249 [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WK7-45F4TXM-2W&_user=4187488&_coverDate=02/04/2000&_rdoc=1&_fmt=high&_orig=search&_origin=search&_sort=d&_docanchor=&view=c&_acct=C000062504&_version=1&_urlVersion=0&_userid=4187488&md5=221a9d3b8b66f6f908a8d93c6b10f18f&searchtype=a#secx12 doi:10.1006/jmbi.1999.3445] </ref>. | ||
| - | == | + | ==Structure== |
| - | The structure of | + | The secondary structure of papain consists of 7 <scene name='Sandbox_30/Papain_secondary_helices/3'>α helices</scene>, 17 <scene name='Sandbox_30/Papain_secondary_sheets/3'>β strands</scene>, all of which are antiparallel, and a large amount (about 50% of total residues) of <scene name='Sandbox_30/Papain_secondary_orf/4'>ordered non-repetitive structures</scene>. The <scene name='Sandbox_30/Papain_rainbow/3'>rainbow coloration view</scene>, which goes from blue (amino terminus) to red (carboxyl terminus) is useful for tracing the order of these structures through the chain. These secondary structures form as a result of favorable hydrogen bonding interactions within the polypeptide backbone. Meanwhile, secondary structures are kept in place by hydrophobic interactions and hydrogen bonds between sidechains of adjacent structures. For example, the <scene name='Sandbox_30/Papain_secondary_helices_1/1'>first helix</scene> (residues 25-42) is maintained as a result of <scene name='Sandbox_30/Papain_secondary_helices_hbond/3'>hydrogen bonds</scene> between backbone carbonyl atoms and the hydrogen on the amide nitrogen four residues away. However, <scene name='Sandbox_30/Papain_secondary_heliceshbond2/1'>no hydrogen bonds</scene> are present between this helix and the rest of the protein, suggesting that this helix is coordinated primarily by hydrophobic interactions. This is reasonable given its central location in the enzyme. As expected, the helix contains many <scene name='Sandbox_30/Papain_secondary_helix1_phobic/1'>hydrophobic residues</scene> (red residues are hydrophilic). The tertiary structure of papain is also maintained by three <scene name='Sandbox_30/Papain_disulfides/3'>disulfide bonds</scene>, which connect <scene name='Sandbox_30/Papain_disulfides_22-63/1'>Cys-22 to Cys63</scene>, |
| - | <scene name='Sandbox_30/ | + | <scene name='Sandbox_30/Papain_disulfides_56-95/1'>Cys-56 to Cys-95</scene>, and <scene name='Sandbox_30/Papain_disulfides_153-200/1'>Cys-153 to Cys-200</scene><ref name="9PAP PDB" />. These disulfide bonds are likely important in conserving the structural integrity of the enzyme as it operates in extracellular environments at high temperatures. |
| - | <scene name='Sandbox_30/ | + | |
| - | + | ||
| + | ===Residue Distribution=== | ||
| + | As can be seen from its <scene name='Sandbox_30/Papain_polarity_ballandstick/3'>ball and stick model</scene>, papain contains a variety of acidic (red), basic (blue), hydrophilic uncharged (pink), and hydrophobic (gray) residues. Since the enzyme operates in free solution, it would be expected that the hydrophilic residues would reside mostly on it exterior, shielding a hydrophobic core. This can be plainly seen from papain's <scene name='Sandbox_30/Papain_polarity_spacefill/4'>space filling model</scene>, in which hydrophilic residues are pink and hydrophobic residues are orange. Upon removal of the hydrophilic residues, the <scene name='Sandbox_30/Papain_hydrophobic_spacefill/3'>hydrophobic core</scene> of the enzyme is easy to see. As with all proteins, it is this hydrophobic segregation that allows the protein to properly fold and maintain a meaningful shape. | ||
===Ligands=== | ===Ligands=== | ||
| - | [[Image: | + | The crystallization procedure used to obtain the 9PAP structure was carried out using a 62% (w/w) methanol and water crystallization medium. Thus, the papain in this crystal structure is coordinated by <scene name='Sandbox_30/Papain_methanol/1'>methanol molecules</scene>, 29 of which are shown, and <scene name='Sandbox_30/Papain_water/1'>water molecules</scene>, 21 of which occur between adjacent papain molecules and are probably important in maintaining their structural integrity<ref name="9PAP PDB" />. Both |
| - | + | <scene name='Sandbox_30/Papain_methanol_h-bondint/1'>methanol</scene> (red and grey) and <scene name='Sandbox_30/Papain_water_h-bonds/3'>water</scene> (purple) molecules form hydrogen bonds with many different residues, which are shown as ball and stick structures in the diagrams. Some of the ethanol molecules also have <scene name='Sandbox_30/Papain_methanol_hydrophobicint/3'>hydrophobic interactions</scene> with the enzyme. | |
| + | |||
| + | |||
| + | ==Catalytic Mechanism== | ||
| + | <applet load='1pop' size='300' frame='true' align='right' scene='Sandbox_30/Papain_ligand_default/4' caption='Papain crystallized with substrate analog leupeptin (blue) covalently bound to the catalytic Cys-25. The PBD code for this structure is 1POP.' /> | ||
| + | [[Image:Papainmech6.jpg|200px|left|thumb| General mechanism of papain catalysis<ref>[http://chemistry.umeche.maine.edu/CHY431/Peptidase10.html] University of Maine</ref>. Arg-175, which orients His 159, and Gln-19, which contributes to the formation of the oxyanion hole, are not shown.]] | ||
| + | Like serine proteases, cysteine proteases contain a <scene name='Sandbox_30/Papain_ligand_active-site/3'>catalytic triad</scene> of residues. In the case of papain, these residues are Cys-25, His-159, and Arg-175. Papain also contains a fourth residue, <scene name='Sandbox_30/Papain_ligand_active-sitegln19/5'>Gln-19</scene> that has been shown to play an important role in catalysis and is likely involved in the formation of the oxyanion hole. The mechanism begins when a peptide binds to the active site. Cys-25 is then deprotonated by His-159 and attacks the substrate carbonyl carbon. This forms a covalent, tetrahedral intermediate that is stabilized by the oxyanion hole. Next, His-159 acts as a general acid, protonating the nitrogen in the peptide bond, which acts as a leaving group as the carbonyl reforms. This now free C-terminal portion of the peptide is released. Water then enters the active site and attacks the carbonyl carbon while it is deprotonated by His-159, again forming an oxyanion hole-stabilized tetradral covalent intermediate. Finally, the carbonyl reforms and the Cys-25 sulfur acts as a leaving group, releasing the N-terminal portion of the peptide and regenerating the enzyme<ref name="Harrison">[http://pubs.acs.org/doi/abs/10.1021/ja9711472]Harrison, M.J., N.A. Burton, and I.H. Hillier. 1997. Catalytic Mechanism of the Enzyme Papain: Predictions with a Hybrid Quantum Mechanical/Molecular Mechanical Potential. J. Am. Chem. Soc. 119: 12285-12291</ref>. | ||
| + | |||
| + | |||
| + | |||
| - | == | + | ===Substrate Binding=== |
| - | + | In order to investigate binding of protein substrates to papain, the enzyme was crystallized with the broad-spectrum competitive protease inhibitor leupeptin, shown in blue in the ribbon diagram. It has the structure Ac-Leu-Leu-Arginal, where Ac is an acetyl group attached to the nitrogen of the first leucine. The inhibitor functions by binding to the enzyme's active site, where the catalytic nucleophile (cysteine in papain) attacks the arginal aldehyde. This forms a tight-binding transition state from which the normal catalytic mechanism cannot proceed, due to this carbonyl having no potential leaving groups bonded to it. Analysis of the resulting structure revealed that the | |
| + | <scene name='Sandbox_30/Papain_inhibitor_activesite/3'>substrate binding pocket</scene> of papain consists primarily of a variety of <scene name='Sandbox_30/Papain_inhibitor_hydrophobics/1'>hydrophobic residues</scene>, including tyrosine, tryptophan, and valine, which coordinate the bound leuptin. Some of the enzyme's residues also make <scene name='Sandbox_30/Papain_inhibitor_h-bonds/2'>hydrogen bonds</scene> with some of the leupeptin atoms. These hydrogen bonds, shown in yellow, include interactions between both hydrogens on both Gln-19 and the amide nitrogen of the catalytic Cys-25 with the arginal carbanion, forming the catalytically important oxyanion hole. In addition, Gly-66 interacts with the second leucine in leupeptin while Asp-158 interacts with a hydrogen on the arginal. These interaction further stabilize and orient the substrate in the binding pocket<ref name="Schroder">[http://www.sciencedirect.com/science/article/pii/001457939381128M] Schröder, E., C. Phillips, E. Garman, K. Harlos, C. Crawford. 1997. X-ray crystallographic structure of a papain-leupeptin complex. FEBS Letters 315: 38-42</ref>. | ||
| - | [[Image:Serine_cleavage.jpg|left|200px]] | ||
| - | <applet scene='Sandbox_30/Big_trypsin_rainbow/1' size='300' frame='true' align='right' caption='Bovine trypsin in complex with UB-THR 10' /> | ||
| - | ==Trypsinogen== | ||
| - | Trypsin's zymogen form is called trypsinogen, and can actually activate itself. Zymogens require a biochemical change to activate. Only an activated form of trypsin can activate the trypsinogen, and this initial activation is carried out by enteropeptidase, which is a serine protease as well. Because activated forms of trypsin can activate others, trypsin is said to be autocatalytic<ref>Voet, Donald et al. Fundamentals of Biochemistry - Life at the Molecular Level. 3rd ed. John Wiley & Sons, Inc. 2008</ref>. In addition to activating itself, it can also activate [http://www.proteopedia.org/wiki/index.php/Chymotrypsin chymotrypsin] and [http://www.proteopedia.org/wiki/index.php/Elastase elastase]. | ||
==References== | ==References== | ||
<references /> | <references /> | ||
Current revision
| Please do NOT make changes to this Sandbox. Sandboxes 30-60 are reserved for use by Biochemistry 410 & 412 at Messiah College taught by Dr. Hannah Tims during Fall 2012 and Spring 2013. |
Contents |
Papain
Note: For some reason, using the "hydrogen bonds" link in the structure section breaks the jmol display and results in scenes after that point having random disulfide bonds and solvent molecules floating in mid-air. I've tried to fix it but to no avail, so if this happens just refresh the page and everything should work again. Oh look, now the new h-bond link in the substrate binding section does the same thing. If you see disembodied floating solvent molecules after clicking it, just refresh the page.
Overview
|
Papain is a 23.4 kDa, 212 residue cysteine endopeptidase originating from the fruit of Carica papaya, where it is present in significant amounts along with three other cysteine proteases, chymopapain, glycyl endopeptidase, and caricain[1][2][3]. Its action was first described by G.C. Roy in 1873. It was studied intensively from the 1950s to the 1960s, during which time it became the second enzyme ever to have its structure determined by x-ray crystallography. Finally, high resolution structural analysis in the 1980s allowed an accurate description of the enzyme's active site[3]. Papain displays very wide hydrolase activity, serving as a general amidase and esterase in addition to its protease activity. As a protease, papain can hydrolyze bonds of basic amino acids, leucine, and glycine. It shows preference for residues preceded by a large hydrophobic residue, but will not cleave if valine is present on the carboxyl side of a potential cleavage site. In addition to being a very non-specific enzyme, papain is also unusually heat resistant, with maximal activity occurring at a temperature of 65° C. These properties have led to use of papain in a large variety areas. One of these areas is biological research, where papain is utilized in cell isolation. It is also useful in immunological techniques because of its ability to cleave the connection between the crystallizable fragment domain and the immunoglobulin domain of antibodies[2]. Additionally, papain has found use outside of research as an inflammation control agent, a digestive aid, and even a meat tenderizer[4].
Structure
The secondary structure of papain consists of 7 , 17 , all of which are antiparallel, and a large amount (about 50% of total residues) of . The , which goes from blue (amino terminus) to red (carboxyl terminus) is useful for tracing the order of these structures through the chain. These secondary structures form as a result of favorable hydrogen bonding interactions within the polypeptide backbone. Meanwhile, secondary structures are kept in place by hydrophobic interactions and hydrogen bonds between sidechains of adjacent structures. For example, the (residues 25-42) is maintained as a result of between backbone carbonyl atoms and the hydrogen on the amide nitrogen four residues away. However, are present between this helix and the rest of the protein, suggesting that this helix is coordinated primarily by hydrophobic interactions. This is reasonable given its central location in the enzyme. As expected, the helix contains many (red residues are hydrophilic). The tertiary structure of papain is also maintained by three , which connect , , and [1]. These disulfide bonds are likely important in conserving the structural integrity of the enzyme as it operates in extracellular environments at high temperatures.
Residue Distribution
As can be seen from its , papain contains a variety of acidic (red), basic (blue), hydrophilic uncharged (pink), and hydrophobic (gray) residues. Since the enzyme operates in free solution, it would be expected that the hydrophilic residues would reside mostly on it exterior, shielding a hydrophobic core. This can be plainly seen from papain's , in which hydrophilic residues are pink and hydrophobic residues are orange. Upon removal of the hydrophilic residues, the of the enzyme is easy to see. As with all proteins, it is this hydrophobic segregation that allows the protein to properly fold and maintain a meaningful shape.
Ligands
The crystallization procedure used to obtain the 9PAP structure was carried out using a 62% (w/w) methanol and water crystallization medium. Thus, the papain in this crystal structure is coordinated by , 29 of which are shown, and , 21 of which occur between adjacent papain molecules and are probably important in maintaining their structural integrity[1]. Both (red and grey) and (purple) molecules form hydrogen bonds with many different residues, which are shown as ball and stick structures in the diagrams. Some of the ethanol molecules also have with the enzyme.
Catalytic Mechanism
|
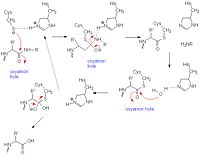
Like serine proteases, cysteine proteases contain a of residues. In the case of papain, these residues are Cys-25, His-159, and Arg-175. Papain also contains a fourth residue, that has been shown to play an important role in catalysis and is likely involved in the formation of the oxyanion hole. The mechanism begins when a peptide binds to the active site. Cys-25 is then deprotonated by His-159 and attacks the substrate carbonyl carbon. This forms a covalent, tetrahedral intermediate that is stabilized by the oxyanion hole. Next, His-159 acts as a general acid, protonating the nitrogen in the peptide bond, which acts as a leaving group as the carbonyl reforms. This now free C-terminal portion of the peptide is released. Water then enters the active site and attacks the carbonyl carbon while it is deprotonated by His-159, again forming an oxyanion hole-stabilized tetradral covalent intermediate. Finally, the carbonyl reforms and the Cys-25 sulfur acts as a leaving group, releasing the N-terminal portion of the peptide and regenerating the enzyme[6].
Substrate Binding
In order to investigate binding of protein substrates to papain, the enzyme was crystallized with the broad-spectrum competitive protease inhibitor leupeptin, shown in blue in the ribbon diagram. It has the structure Ac-Leu-Leu-Arginal, where Ac is an acetyl group attached to the nitrogen of the first leucine. The inhibitor functions by binding to the enzyme's active site, where the catalytic nucleophile (cysteine in papain) attacks the arginal aldehyde. This forms a tight-binding transition state from which the normal catalytic mechanism cannot proceed, due to this carbonyl having no potential leaving groups bonded to it. Analysis of the resulting structure revealed that the of papain consists primarily of a variety of , including tyrosine, tryptophan, and valine, which coordinate the bound leuptin. Some of the enzyme's residues also make with some of the leupeptin atoms. These hydrogen bonds, shown in yellow, include interactions between both hydrogens on both Gln-19 and the amide nitrogen of the catalytic Cys-25 with the arginal carbanion, forming the catalytically important oxyanion hole. In addition, Gly-66 interacts with the second leucine in leupeptin while Asp-158 interacts with a hydrogen on the arginal. These interaction further stabilize and orient the substrate in the binding pocket[7].
References
- ↑ 1.0 1.1 1.2 [1] 9PAP PDB
- ↑ 2.0 2.1 [2] Sigma Aldrich
- ↑ 3.0 3.1 [3] Worthington Biochemical Corporation
- ↑ [4] WebMD
- ↑ [5] University of Maine
- ↑ [6]Harrison, M.J., N.A. Burton, and I.H. Hillier. 1997. Catalytic Mechanism of the Enzyme Papain: Predictions with a Hybrid Quantum Mechanical/Molecular Mechanical Potential. J. Am. Chem. Soc. 119: 12285-12291
- ↑ [7] Schröder, E., C. Phillips, E. Garman, K. Harlos, C. Crawford. 1997. X-ray crystallographic structure of a papain-leupeptin complex. FEBS Letters 315: 38-42