Guanine-Binding Riboswitch
From Proteopedia
(Difference between revisions)
m |
|||
| (5 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | [[ | + | <StructureSection load='4fe5' size='350' side='right' caption='Crystal structure of the xpt-pbuX guanine riboswitch aptamer domain in complex with hypoxanthine (PDB entry [[4fe5]])' scene=''> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==What is a Riboswitch?== | ==What is a Riboswitch?== | ||
| Line 22: | Line 17: | ||
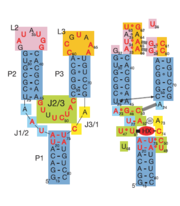
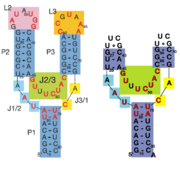
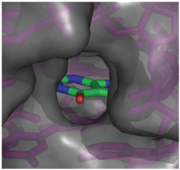
| - | + | When the riboswitch is in a ligand-bound state, the RNA to forms a three-dimentional fold in which the terminal loops <scene name='Guanine-Binding_Riboswitch/Terminal_loops/1'>L2 and L3</scene> become connected through a series of hydrogen bonds (Fig. 1). This causes the <scene name='Guanine-Binding_Riboswitch/Helix_p2/2'>P2</scene> and <scene name='Guanine-Binding_Riboswitch/Helix_p3/2'>P3</scene> helices to become parallel to each other. The unfavorable electrostatic interactions of the parallel ribose-phosphate backbones are neutralized through the binding multiple Co(NH<sub>3</sub>)<sub>6</sub><sup>3+</sup> <scene name='Guanine-Binding_Riboswitch/Cobalt_cations/2'>cations</scene> between the backbones. The interactions between the terminal loops stabilize the tertiary structure of the riboswitch, and are essential for ligand binding to occur. Experiments in which the loops are replaced with extremely stable UUCG sequences (Fig. 2) have demonstrated that alterations to the terminal loops prevent the loop-loop interactions from forming, and eliminate the riboswitch's ability to recognize hypoxanthine as a result.[[Image:Riboswitch_loops_replaced.png|thumb|'''Fig. 2:''' Left: Riboswitch with wild-type terminal loops. Right: Riboswitch with terminal loops replaced by UUCG tetraloops.]] As such, this tertiary interaction is required for ligand recognition, even though it never contacts the ligand directly<ref name=bateyetal/>. | |
| Line 30: | Line 25: | ||
==Gene Regulation== | ==Gene Regulation== | ||
| - | This riboswitch acts as a transcriptional regulation mechanism in the operon of the ''xpt-pbuX'' purine salvaging gene of ''B. subtilis,'' and directly represses gene expression in response to an increase in intracellular concentrations of guanine, xanthine, or hypoxanthine. Transcription of the initial ~90 nucleotides of the sequence triggers the interaction of the <scene name='Guanine-Binding_Riboswitch/Terminal_loops/1'>L2 and L3</scene> loops. This results in the organization of the | + | This riboswitch acts as a transcriptional regulation mechanism in the operon of the ''xpt-pbuX'' purine salvaging gene of ''B. subtilis,'' and directly represses gene expression in response to an increase in intracellular concentrations of guanine, xanthine, or hypoxanthine. Transcription of the initial ~90 nucleotides of the sequence triggers the interaction of the <scene name='Guanine-Binding_Riboswitch/Terminal_loops/1'>L2 and L3</scene> loops. This results in the organization of the <scene name='Guanine-Binding_Riboswitch/Binding_junction/4'>three-way junction binding pocket</scene> such that it is partially unstructured to allow access to the site while still allowing efficient ligand binding. |
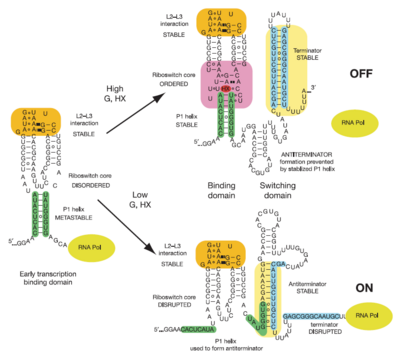
Illustrated below are the conformational changes made by the riboswitch in response to changes in ligand concentration. At low concentrations of ligand (in this case hypoxanthine), the <scene name='Guanine-Binding_Riboswitch/Helix_p1/4'>P1 helix</scene> becomes destabilized. This allows the RNA to incorporate part of the helix into an antiterminator element as illustrated below, resulting in continued transcription. At sufficiently high concentrations of ligand, the nucleobase binds to the junction and stabilizes the <scene name='Guanine-Binding_Riboswitch/Helix_p1/4'>P1 helix</scene>. This prevents the helix from being incorporated into an antiterminator element, resulting in the formation of a stable stem-loop structure that halts transcription via Rho-independent termination<ref name=bateyetal/>. | Illustrated below are the conformational changes made by the riboswitch in response to changes in ligand concentration. At low concentrations of ligand (in this case hypoxanthine), the <scene name='Guanine-Binding_Riboswitch/Helix_p1/4'>P1 helix</scene> becomes destabilized. This allows the RNA to incorporate part of the helix into an antiterminator element as illustrated below, resulting in continued transcription. At sufficiently high concentrations of ligand, the nucleobase binds to the junction and stabilizes the <scene name='Guanine-Binding_Riboswitch/Helix_p1/4'>P1 helix</scene>. This prevents the helix from being incorporated into an antiterminator element, resulting in the formation of a stable stem-loop structure that halts transcription via Rho-independent termination<ref name=bateyetal/>. | ||
| - | [[Image:Function_of_riboswitch.png]] | + | [[Image:Function_of_riboswitch.png|400px]] |
==Potential Applications of Riboswitches== | ==Potential Applications of Riboswitches== | ||
| Line 43: | Line 38: | ||
Artificial riboswitches have also been engineered for the manipulation of gene expression. Identifying the principles of riboswitch-mediated regulation may lead to the development of engineered ligands capable of modulating gene expression. More detailed characterization of the distribution and function of riboswitches across and within different genomes is essential to determine their precise role as riboregulators and potential drug targets<ref name=singhetal/>. | Artificial riboswitches have also been engineered for the manipulation of gene expression. Identifying the principles of riboswitch-mediated regulation may lead to the development of engineered ligands capable of modulating gene expression. More detailed characterization of the distribution and function of riboswitches across and within different genomes is essential to determine their precise role as riboregulators and potential drug targets<ref name=singhetal/>. | ||
| + | ==3D structures of riboswitch== | ||
| + | |||
| + | [[Riboswitch]] | ||
| + | |||
| + | </StructureSection> | ||
==Reference== | ==Reference== | ||
Current revision
| |||||||||||
Reference
- ↑ 1.0 1.1 1.2 1.3 Singh P, Bandyopadhyay P, Bhattacharya S, Krishnamachari A, Sengupta S. Riboswitch detection using profile hidden Markov models. BMC Bioinformatics. 2009 Oct 8;10:325. PMID:19814811 doi:10.1186/1471-2105-10-325
- ↑ 2.0 2.1 Breaker, Ronald R. (28 March, 2008). Complex Riboswitches. Science, 319(5871), 1795-1797. doi:10.1126/science.1152621
- ↑ 3.0 3.1 3.2 3.3 3.4 Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004 Nov 18;432(7015):411-5. PMID:15549109 doi:10.1038/nature03037