Hemagglutinin
From Proteopedia
(Difference between revisions)
m |
|||
| (46 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
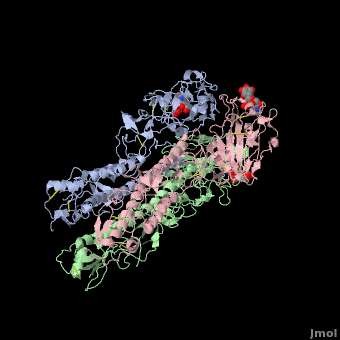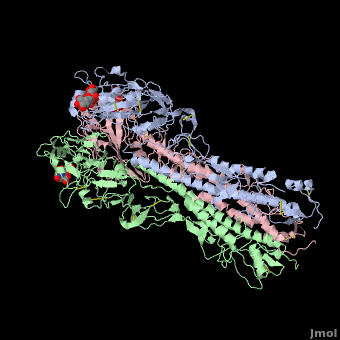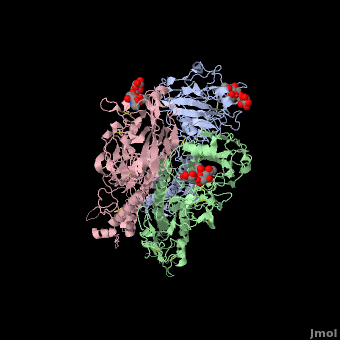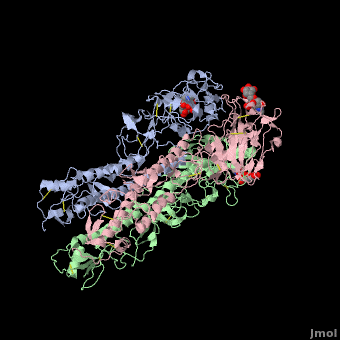
| - | [[ | + | <StructureSection load='' size='350' side='right' scene ='Hemagglutinin/Blackground2wre/1' caption='Structure of glycosylated viral hemagglutinin trimer complex with galactose ([[2wre]])'> |
| - | + | == Function == | |
| - | [[Hemagglutinin|Hemagglutinins]] (HA) are one of the two antigenic Glycoproteins inserted into the influenza virus' membrane. There are at least 16 different forms of HA | + | [[Hemagglutinin|Hemagglutinins]] (HA) are one of the two antigenic Glycoproteins inserted into the influenza virus' membrane. There are at least 16 different forms of HA antigens classified from H1 to H16. H1, H2, H3 are specific to human [[influenza]]. HAs have two main functions crucial for the [[Viral Infections|viral infection]] cycle: |
| - | 1. On the target cell, HA | + | 1. On the target cell, HA binds the receptor on the cell membrane which is contains sialic acid <ref>Gottschalk A. Chemistry of virus receptors. The Viruses. 1959;3:51–61.</ref> allowing the virus/cell interaction. |
2. HA induce the fusion between the host cell and the virus which can entry into the cytoplasm. | 2. HA induce the fusion between the host cell and the virus which can entry into the cytoplasm. | ||
| - | Because the HA are the major Antigens of the Virus, Antibodies recognized them and for this reason HA often change. | + | Because the HA are the major Antigens of the Virus, [[Antibody|Antibodies]] recognized them and for this reason HA often change. |
| + | * '''Dr hemagglutinin''' is found in urinary tract pathogenic ''E. coli'' and recognizes Dr blood group antigen<ref>PMID: 3054548</ref>. | ||
| + | * '''Phytohemagglutinin''' is found in bean seeds and has sugar binding and hemagglutinin functions<ref>PMID: 8702788</ref>. | ||
| + | * '''Hemagglutinin-neuraminidase''' (HN) is multifunctional. It possesses both the receptor recognition and neuraminidase activities<ref>PMID:21680512</ref>. For details see [[Mumps Virus Hemagglutinin Neuraminidase Protein]]. | ||
| + | |||
| + | For discussion of influenza hemagglutinin see<br /> | ||
| + | *[[Influenza hemagglutinin]]<br /> | ||
| + | *[[Influenza]]<br /> | ||
| + | *[[User:Michael Strong/H1N1/HA]]<br /> | ||
| + | *[[User:Michael Strong/H1N1/HA/MSA]] for multiple sequence alignment. | ||
==Hemagglutinin Structure== | ==Hemagglutinin Structure== | ||
| - | HA is an <scene name='Hemagglutinin/Homotrimer/2'>homotrimer</scene> integral membrane glycoprotein. Each monomer is synthesized like a single polypeptide chain almost 550 amino acids. This precursor is then glycosilated and cleaved into two smaller polypeptides by removal of | + | HA is an <scene name='Hemagglutinin/Homotrimer/2'>homotrimer</scene> integral membrane glycoprotein. Each monomer is synthesized like a single polypeptide chain almost 550 amino acids. This precursor is then glycosilated and cleaved into two smaller polypeptides by removal of Arginine 329; at the same time, a conformational change occurs in the monomer. |
The HA1 and HA2 subunits are covalently attached by a <scene name='SAndbox_159/Disulfide_bond/2'>disulfide bond</scene> from HA1 position 14 to HA2 position 467(*). These two chains form one monomer, and the noncovalentely association of three monomers forms one hemagglutinin molecule: (HA1+HA2)3. It is principally stabilized by packing of the alpha-helixes. All molecules are 135 Angström long. | The HA1 and HA2 subunits are covalently attached by a <scene name='SAndbox_159/Disulfide_bond/2'>disulfide bond</scene> from HA1 position 14 to HA2 position 467(*). These two chains form one monomer, and the noncovalentely association of three monomers forms one hemagglutinin molecule: (HA1+HA2)3. It is principally stabilized by packing of the alpha-helixes. All molecules are 135 Angström long. | ||
| Line 16: | Line 25: | ||
The <scene name='SAndbox_159/Ha2/2'>HA2 subunit</scene> (221 amino acids) has a hair spin structure composed by two antiparallel alpha-helixes. One of these belongs to the longest alpha-helixes in globular protein: it is 50 angstrom long. The hydrophobic N-terminus part of HA2, called fusion peptide, is close to a protease’s cleavage site: this protease is implied in the virus’ entry in the host cell. | The <scene name='SAndbox_159/Ha2/2'>HA2 subunit</scene> (221 amino acids) has a hair spin structure composed by two antiparallel alpha-helixes. One of these belongs to the longest alpha-helixes in globular protein: it is 50 angstrom long. The hydrophobic N-terminus part of HA2, called fusion peptide, is close to a protease’s cleavage site: this protease is implied in the virus’ entry in the host cell. | ||
| - | HA1 and HA2 are bound each other by a disulfide bind. The three long alpha-helixes (of the three HA2) are coiled-coil to form a central region of 40 Angström, and thank to the hydrophobic amino acids and those which form salt bridges bound, we obtain a HA stabilized. | + | HA1 and HA2 are bound each other by a disulfide bind. The three long alpha-helixes (of the three HA2) are coiled-coil to form a central region of 40 Angström, and thank to the hydrophobic amino acids and those which form salt bridges bound, we obtain a HA stabilized.<ref>PMID: 3304138</ref> |
==2WRE== | ==2WRE== | ||
| Line 34: | Line 43: | ||
- the last is named 190-helix | - the last is named 190-helix | ||
| - | [[Image:Avian.jpg|300px|thumb]] | ||
| + | [[Image:Avian.jpg|300px|thumb]] | ||
| + | {{Clear}} | ||
The residues of these loops 130 and 220 have carbonyl oxygens and amide nitrogens of peptide bonds exposed with potential to interact with the receptor, which is composed by Gal-1 and Sia-2. | The residues of these loops 130 and 220 have carbonyl oxygens and amide nitrogens of peptide bonds exposed with potential to interact with the receptor, which is composed by Gal-1 and Sia-2. | ||
The avian H2 can bind the human receptor because of intramolecular Hydrogen bond network created by Gln 226 and by Asn 186. | The avian H2 can bind the human receptor because of intramolecular Hydrogen bond network created by Gln 226 and by Asn 186. | ||
| Line 41: | Line 51: | ||
<scene name='SAndbox_159/Amino_acids_in_the_stability/1'>(Cys 97, Pro 99, Cis139, Pro 147, Tyr 195, Arg 229).</scene>(*) | <scene name='SAndbox_159/Amino_acids_in_the_stability/1'>(Cys 97, Pro 99, Cis139, Pro 147, Tyr 195, Arg 229).</scene>(*) | ||
The Sia-1-Gal-2 glycosidic bond adopts a cis conformation. Extensive hydrogen bond direction are settled between avian and Gal-2 of the receptor: two water molecules (Wat-1 and Wat-2) have a significant role in mediating these interactions(see fig.1). | The Sia-1-Gal-2 glycosidic bond adopts a cis conformation. Extensive hydrogen bond direction are settled between avian and Gal-2 of the receptor: two water molecules (Wat-1 and Wat-2) have a significant role in mediating these interactions(see fig.1). | ||
| - | The site chain of Lys-222 and the main chain carbonyl at 225 are linked by Wat-1 to the 3'OH of Gal-2. Gln-226 and Asn 186 form hydrogen bonds with the hydroxyl groups of 4'C of Gal-2 and 9'C of Sia-1. | + | The site chain of Lys-222 and the main chain carbonyl at 225 are linked by Wat-1 to the 3'OH of Gal-2. Gln-226 and Asn 186 form hydrogen bonds with the hydroxyl groups of 4'C of Gal-2 and 9'C of Sia-1.<ref>Cell Binding protein in Avian Influenza; Jack Cerchiara,'06 and Brendan Holsberry, 07;http://biology.kenyon.edu/BMB/Chime2/2005/Cerchiara-Holsberry/FRAMES/start.htm</ref> |
==Oligosaccharides== | ==Oligosaccharides== | ||
| - | Some <scene name='SAndbox_159/Asparagines/1'>Asparagines</scene> of HA1 (8, 22, 38, 81, 165, 285)(*) have oligosaccharides chains attached to them. The seventh oligasaccharide is linked to an Aspargine of HA2 (484). All of them are complex oligosaccharides and are on the lateral surfaces of the molecule, except for site and 165. No precise functions have been assigned to them, even if the oligosaccharide at 165 seems to allow the stabilisation of oligomeric contact between globular units at the top of the protein's structure. | + | Some <scene name='SAndbox_159/Asparagines/1'>Asparagines</scene> of HA1 (8, 22, 38, 81, 165, 285)(*) have oligosaccharides chains attached to them. The seventh oligasaccharide is linked to an Aspargine of HA2 (484). All of them are complex oligosaccharides and are on the lateral surfaces of the molecule, except for site and 165. No precise functions have been assigned to them, even if the oligosaccharide at 165 seems to allow the stabilisation of oligomeric contact between globular units at the top of the protein's structure. |
==Antigenic variability== | ==Antigenic variability== | ||
| Line 52: | Line 62: | ||
The substitutions of amino acids in these sites allow the influenza virus to escape from the immune system and to spread. Major changes in these antigenic regions can produce lethal influenza pandemics, like in 1957. | The substitutions of amino acids in these sites allow the influenza virus to escape from the immune system and to spread. Major changes in these antigenic regions can produce lethal influenza pandemics, like in 1957. | ||
| + | |||
| + | ==How Influenza Escapes Vaccines== | ||
| + | |||
| + | Influenza hemagglutinin (''e.g.'' [[1hgf]]) is a glycoprotein on the surface of [http://en.wikipedia.org/wiki/Influenza_virus influenza virus] particles that enables them to attach to and infect host cells. [[Antibody|Antibodies]] that bind to hemagglutinin are a major defense mechanism that prevent infection. The RNA genome of influenza is characterized by a high mutation rate. Mutations on the surface of hemagglutinin tend to be protective for the virus. They tend to be retained because they tend to reduce the binding strength, and hence the host defensive capability, of antibodies that recognize the un-mutated hemagglutinin<ref name="skehel_review">PMID: 16925526</ref>. Influenza vaccines include hemagglutinins and they induce anti-hemagglutinin antibodies in vaccinated individuals. Often, however, by the time the vaccines can be designed, produced, and disseminated, mutant influenza viruses have arisen that can cause disease in vaccinated individuals<ref name="fluwikipedia">See [http://en.wikipedia.org/wiki/Influenza Influenza] in Wikipedia.</ref>. | ||
| + | If you '''show''' the '''Evolutionary Conservation''' of the structure on this page (using the blue bars below the molecule), you will see highly variable surface amino acids. These represent sites of mutations that have been retained in wild strains of influenza because they improve virus survival<ref name="skehel_review" />. (Added by [[User:Eric Martz|Eric Martz]]). | ||
==Notes== | ==Notes== | ||
| Line 58: | Line 73: | ||
(**) The amino acid 153 lacks for the A monomer chain. | (**) The amino acid 153 lacks for the A monomer chain. | ||
| - | == | + | ==3D structures of hemagglutinin== |
| - | + | [[Hemagglutinin 3D structures]] | |
| - | + | ||
| - | == | + | ==3D Printed Physical Models of Hemagglutinin== |
| - | + | ||
| - | + | Shown below are 3D printed physical models of Hemagglutinin. The model is shown as an alpha carbon backbone, with key sidechains and domains highlighted. It has been designed with precisely embedded magnets that allow the three chains to pull apart into individual pieces. | |
| + | |||
| - | + | [[Image:hemagglutinin_1_centerForBiomolecularModeling.jpg | 450px]][[Image:hemagglutinin_2_centerForBiomolecularModeling.jpg | 450px]] | |
| - | + | ||
| + | ====The MSOE Center for BioMolecular Modeling==== | ||
| + | |||
| + | [[Image:CbmUniversityLogo.jpg | left | 100px]] | ||
| + | |||
| + | The [http://cbm.msoe.edu MSOE Center for BioMolecular Modeling] uses 3D printing technology to create physical models of protein and molecular structures, making the invisible molecular world more tangible and comprehensible. To view more protein structure models, visit our [http://cbm.msoe.edu/educationalmedia/modelgallery/ Model Gallery]. | ||
| + | |||
| + | </StructureSection> | ||
| + | |||
| + | ==Additional Resources== | ||
| + | For additional information, see: [[Influenza]] | ||
| + | <br /> | ||
| + | ==References== | ||
<references/> | <references/> | ||
| - | + | [[Category: Topic Page]] | |
| + | [[Category:3D printer files]] | ||
Current revision
| |||||||||||
Additional Resources
For additional information, see: Influenza
References
- ↑ Gottschalk A. Chemistry of virus receptors. The Viruses. 1959;3:51–61.
- ↑ Nowicki B, Hull R, Moulds J. Use of the Dr hemagglutinin of uropathogenic Escherichia coli to differentiate normal from abnormal red cells in paroxysmal nocturnal hemoglobinuria. N Engl J Med. 1988 Nov 10;319(19):1289-90. PMID:3054548 doi:10.1056/NEJM198811103191916
- ↑ Hamelryck TW, Dao-Thi MH, Poortmans F, Chrispeels MJ, Wyns L, Loris R. The crystallographic structure of phytohemagglutinin-L. J Biol Chem. 1996 Aug 23;271(34):20479-85. PMID:8702788
- ↑ Kim SH, Subbiah M, Samuel AS, Collins PL, Samal SK. Roles of the fusion and hemagglutinin-neuraminidase proteins in replication, tropism, and pathogenicity of avian paramyxoviruses. J Virol. 2011 Sep;85(17):8582-96. doi: 10.1128/JVI.00652-11. Epub 2011 Jun 15. PMID:21680512 doi:http://dx.doi.org/10.1128/JVI.00652-11
- ↑ Wiley DC, Skehel JJ. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365-94. PMID:3304138 doi:http://dx.doi.org/10.1146/annurev.bi.56.070187.002053
- ↑ Liu J, Stevens DJ, Haire LF, Walker PA, Coombs PJ, Russell RJ, Gamblin SJ, Skehel JJ. Structures of receptor complexes formed by hemagglutinins from the Asian Influenza pandemic of 1957. Proc Natl Acad Sci U S A. 2009 Oct 6;106(40):17175-80. Epub 2009 Sep 28. PMID:19805083
- ↑ Cell Binding protein in Avian Influenza; Jack Cerchiara,'06 and Brendan Holsberry, 07;http://biology.kenyon.edu/BMB/Chime2/2005/Cerchiara-Holsberry/FRAMES/start.htm
- ↑ 8.0 8.1 Knossow M, Skehel JJ. Variation and infectivity neutralization in influenza. Immunology. 2006 Sep;119(1):1-7. PMID:16925526 doi:http://dx.doi.org/10.1111/j.1365-2567.2006.02421.x
- ↑ See Influenza in Wikipedia.
Proteopedia Page Contributors and Editors (what is this?)
Julien Madouasse, Michal Harel, David Canner, Mark Hoelzer, Ann Taylor, Alexander Berchansky, Joel L. Sussman, Marius Mihasan