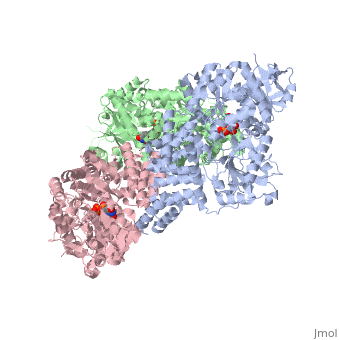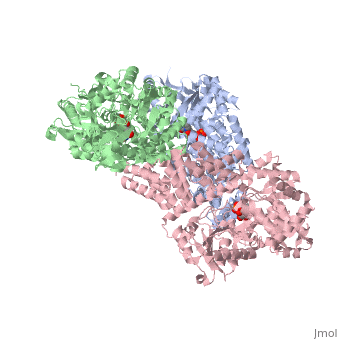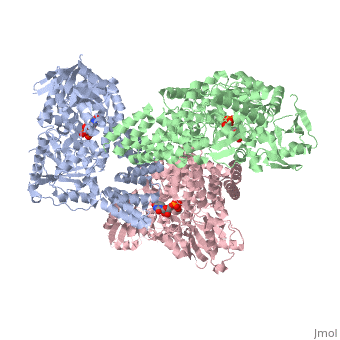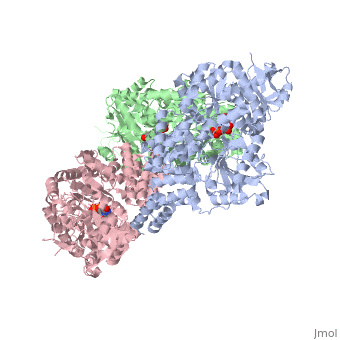
| Structural highlights
Disease
OGT1_HUMAN Regulation of OGT activity and altered O-GlcNAcylations are implicated in diabetes and Alzheimer disease. O-GlcNAcylation of AKT1 affects insulin signaling and, possibly diabetes. Reduced O-GlcNAcylations and resulting increased phosphorylations of MAPT/TAU are observed in Alzheimer disease (AD) brain cerebrum.
Function
OGT1_HUMAN Catalyzes the transfer of a single N-acetylglucosamine from UDP-GlcNAc to a serine or threonine residue in cytoplasmic and nuclear proteins resulting in their modification with a beta-linked N-acetylglucosamine (O-GlcNAc). Glycosylates a large and diverse number of proteins including histone H2B, AKT1, PFKL, KMT2E/MLL5, MAPT/TAU and HCFC1. Can regulate their cellular processes via cross-talk between glycosylation and phosphorylation or by affecting proteolytic processing. Involved in insulin resistance in muscle and adipocyte cells via glycosylating insulin signaling components and inhibiting the 'Thr-308' phosphorylation of AKT1, enhancing IRS1 phosphorylation and attenuating insulin signaling. Involved in glycolysis regulation by mediating glycosylation of 6-phosphofructokinase PFKL, inhibiting its activity. Component of a THAP1/THAP3-HCFC1-OGT complex that is required for the regulation of the transcriptional activity of RRM1. Plays a key role in chromatin structure by mediating O-GlcNAcylation of 'Ser-112' of histone H2B: recruited to CpG-rich transcription start sites of active genes via its interaction with TET proteins (TET1, TET2 or TET3). As part of the NSL complex indirectly involved in acetylation of nucleosomal histone H4 on several lysine residues.[1] [2] [3] [4] [5] [6] [7] [8] [9] [10] [11] [12] [13] [14] [15] Isoform 2: the mitochondrial isoform (mOGT) is cytotoxic and triggers apoptosis in several cell types including INS1, an insulinoma cell line.[16] [17] [18] [19] [20] [21] [22] [23] [24] [25] [26] [27] [28] [29] [30]
See Also
References
- ↑ Yang X, Zhang F, Kudlow JE. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell. 2002 Jul 12;110(1):69-80. PMID:12150998
- ↑ Yang X, Ongusaha PP, Miles PD, Havstad JC, Zhang F, So WV, Kudlow JE, Michell RH, Olefsky JM, Field SJ, Evans RM. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature. 2008 Feb 21;451(7181):964-9. PMID:18288188 doi:10.1038/nature06668
- ↑ Liu F, Shi J, Tanimukai H, Gu J, Gu J, Grundke-Iqbal I, Iqbal K, Gong CX. Reduced O-GlcNAcylation links lower brain glucose metabolism and tau pathology in Alzheimer's disease. Brain. 2009 Jul;132(Pt 7):1820-32. doi: 10.1093/brain/awp099. Epub 2009 May 18. PMID:19451179 doi:10.1093/brain/awp099
- ↑ Fujiki R, Chikanishi T, Hashiba W, Ito H, Takada I, Roeder RG, Kitagawa H, Kato S. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature. 2009 May 21;459(7245):455-9. Epub 2009 Apr 19. PMID:19377461 doi:nature07954
- ↑ Cai Y, Jin J, Swanson SK, Cole MD, Choi SH, Florens L, Washburn MP, Conaway JW, Conaway RC. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J Biol Chem. 2010 Feb 12;285(7):4268-72. doi: 10.1074/jbc.C109.087981. Epub 2009 , Dec 14. PMID:20018852 doi:10.1074/jbc.C109.087981
- ↑ Whelan SA, Dias WB, Thiruneelakantapillai L, Lane MD, Hart GW. Regulation of insulin receptor substrate 1 (IRS-1)/AKT kinase-mediated insulin signaling by O-Linked beta-N-acetylglucosamine in 3T3-L1 adipocytes. J Biol Chem. 2010 Feb 19;285(8):5204-11. doi: 10.1074/jbc.M109.077818. Epub 2009 , Dec 17. PMID:20018868 doi:http://dx.doi.org/10.1074/jbc.M109.077818
- ↑ Mazars R, Gonzalez-de-Peredo A, Cayrol C, Lavigne AC, Vogel JL, Ortega N, Lacroix C, Gautier V, Huet G, Ray A, Monsarrat B, Kristie TM, Girard JP. The THAP-zinc finger protein THAP1 associates with coactivator HCF-1 and O-GlcNAc transferase: a link between DYT6 and DYT3 dystonias. J Biol Chem. 2010 Apr 30;285(18):13364-71. doi: 10.1074/jbc.M109.072579. Epub, 2010 Mar 3. PMID:20200153 doi:10.1074/jbc.M109.072579
- ↑ Shin SH, Love DC, Hanover JA. Elevated O-GlcNAc-dependent signaling through inducible mOGT expression selectively triggers apoptosis. Amino Acids. 2011 Mar;40(3):885-93. doi: 10.1007/s00726-010-0719-8. Epub 2010 Sep, 8. PMID:20824293 doi:http://dx.doi.org/10.1007/s00726-010-0719-8
- ↑ Daou S, Mashtalir N, Hammond-Martel I, Pak H, Yu H, Sui G, Vogel JL, Kristie TM, Affar el B. Crosstalk between O-GlcNAcylation and proteolytic cleavage regulates the host cell factor-1 maturation pathway. Proc Natl Acad Sci U S A. 2011 Feb 15;108(7):2747-52. doi:, 10.1073/pnas.1013822108. Epub 2011 Feb 1. PMID:21285374 doi:http://dx.doi.org/10.1073/pnas.1013822108
- ↑ Fujiki R, Hashiba W, Sekine H, Yokoyama A, Chikanishi T, Ito S, Imai Y, Kim J, He HH, Igarashi K, Kanno J, Ohtake F, Kitagawa H, Roeder RG, Brown M, Kato S. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature. 2011 Nov 27;480(7378):557-60. doi: 10.1038/nature10656. PMID:22121020 doi:http://dx.doi.org/10.1038/nature10656
- ↑ Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard WA 3rd, Peters EC, Driggers EM, Hsieh-Wilson LC. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science. 2012 Aug 24;337(6097):975-80. doi: 10.1126/science.1222278. PMID:22923583 doi:http://dx.doi.org/10.1126/science.1222278
- ↑ Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, Shih AH, Levine RL, Bernard O, Mercher T, Solary E, Urh M, Daniels DL, Fuks F. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013 Mar 6;32(5):645-55. doi: 10.1038/emboj.2012.357. Epub 2013 Jan 25. PMID:23353889 doi:http://dx.doi.org/10.1038/emboj.2012.357
- ↑ Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013 Jan 24;493(7433):561-4. doi: 10.1038/nature11742. Epub 2012 Dec 9. PMID:23222540 doi:http://dx.doi.org/10.1038/nature11742
- ↑ Jinek M, Rehwinkel J, Lazarus BD, Izaurralde E, Hanover JA, Conti E. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat Struct Mol Biol. 2004 Oct;11(10):1001-7. Epub 2004 Sep 12. PMID:15361863 doi:10.1038/nsmb833
- ↑ Lazarus MB, Nam Y, Jiang J, Sliz P, Walker S. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature. 2011 Jan 27;469(7331):564-7. Epub 2011 Jan 16. PMID:21240259 doi:10.1038/nature09638
- ↑ Yang X, Zhang F, Kudlow JE. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell. 2002 Jul 12;110(1):69-80. PMID:12150998
- ↑ Yang X, Ongusaha PP, Miles PD, Havstad JC, Zhang F, So WV, Kudlow JE, Michell RH, Olefsky JM, Field SJ, Evans RM. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature. 2008 Feb 21;451(7181):964-9. PMID:18288188 doi:10.1038/nature06668
- ↑ Liu F, Shi J, Tanimukai H, Gu J, Gu J, Grundke-Iqbal I, Iqbal K, Gong CX. Reduced O-GlcNAcylation links lower brain glucose metabolism and tau pathology in Alzheimer's disease. Brain. 2009 Jul;132(Pt 7):1820-32. doi: 10.1093/brain/awp099. Epub 2009 May 18. PMID:19451179 doi:10.1093/brain/awp099
- ↑ Fujiki R, Chikanishi T, Hashiba W, Ito H, Takada I, Roeder RG, Kitagawa H, Kato S. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature. 2009 May 21;459(7245):455-9. Epub 2009 Apr 19. PMID:19377461 doi:nature07954
- ↑ Cai Y, Jin J, Swanson SK, Cole MD, Choi SH, Florens L, Washburn MP, Conaway JW, Conaway RC. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J Biol Chem. 2010 Feb 12;285(7):4268-72. doi: 10.1074/jbc.C109.087981. Epub 2009 , Dec 14. PMID:20018852 doi:10.1074/jbc.C109.087981
- ↑ Whelan SA, Dias WB, Thiruneelakantapillai L, Lane MD, Hart GW. Regulation of insulin receptor substrate 1 (IRS-1)/AKT kinase-mediated insulin signaling by O-Linked beta-N-acetylglucosamine in 3T3-L1 adipocytes. J Biol Chem. 2010 Feb 19;285(8):5204-11. doi: 10.1074/jbc.M109.077818. Epub 2009 , Dec 17. PMID:20018868 doi:http://dx.doi.org/10.1074/jbc.M109.077818
- ↑ Mazars R, Gonzalez-de-Peredo A, Cayrol C, Lavigne AC, Vogel JL, Ortega N, Lacroix C, Gautier V, Huet G, Ray A, Monsarrat B, Kristie TM, Girard JP. The THAP-zinc finger protein THAP1 associates with coactivator HCF-1 and O-GlcNAc transferase: a link between DYT6 and DYT3 dystonias. J Biol Chem. 2010 Apr 30;285(18):13364-71. doi: 10.1074/jbc.M109.072579. Epub, 2010 Mar 3. PMID:20200153 doi:10.1074/jbc.M109.072579
- ↑ Shin SH, Love DC, Hanover JA. Elevated O-GlcNAc-dependent signaling through inducible mOGT expression selectively triggers apoptosis. Amino Acids. 2011 Mar;40(3):885-93. doi: 10.1007/s00726-010-0719-8. Epub 2010 Sep, 8. PMID:20824293 doi:http://dx.doi.org/10.1007/s00726-010-0719-8
- ↑ Daou S, Mashtalir N, Hammond-Martel I, Pak H, Yu H, Sui G, Vogel JL, Kristie TM, Affar el B. Crosstalk between O-GlcNAcylation and proteolytic cleavage regulates the host cell factor-1 maturation pathway. Proc Natl Acad Sci U S A. 2011 Feb 15;108(7):2747-52. doi:, 10.1073/pnas.1013822108. Epub 2011 Feb 1. PMID:21285374 doi:http://dx.doi.org/10.1073/pnas.1013822108
- ↑ Fujiki R, Hashiba W, Sekine H, Yokoyama A, Chikanishi T, Ito S, Imai Y, Kim J, He HH, Igarashi K, Kanno J, Ohtake F, Kitagawa H, Roeder RG, Brown M, Kato S. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature. 2011 Nov 27;480(7378):557-60. doi: 10.1038/nature10656. PMID:22121020 doi:http://dx.doi.org/10.1038/nature10656
- ↑ Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard WA 3rd, Peters EC, Driggers EM, Hsieh-Wilson LC. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science. 2012 Aug 24;337(6097):975-80. doi: 10.1126/science.1222278. PMID:22923583 doi:http://dx.doi.org/10.1126/science.1222278
- ↑ Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, Shih AH, Levine RL, Bernard O, Mercher T, Solary E, Urh M, Daniels DL, Fuks F. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013 Mar 6;32(5):645-55. doi: 10.1038/emboj.2012.357. Epub 2013 Jan 25. PMID:23353889 doi:http://dx.doi.org/10.1038/emboj.2012.357
- ↑ Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013 Jan 24;493(7433):561-4. doi: 10.1038/nature11742. Epub 2012 Dec 9. PMID:23222540 doi:http://dx.doi.org/10.1038/nature11742
- ↑ Jinek M, Rehwinkel J, Lazarus BD, Izaurralde E, Hanover JA, Conti E. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat Struct Mol Biol. 2004 Oct;11(10):1001-7. Epub 2004 Sep 12. PMID:15361863 doi:10.1038/nsmb833
- ↑ Lazarus MB, Nam Y, Jiang J, Sliz P, Walker S. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature. 2011 Jan 27;469(7331):564-7. Epub 2011 Jan 16. PMID:21240259 doi:10.1038/nature09638
|