Ann Taylor 115
From Proteopedia
(Difference between revisions)
| (3 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
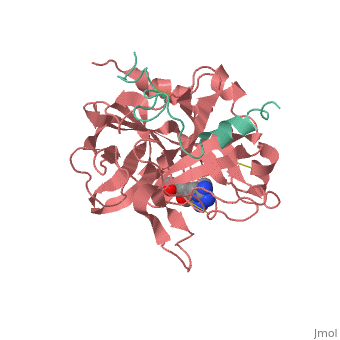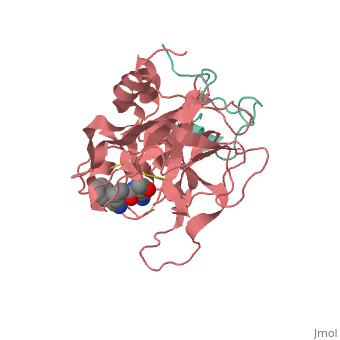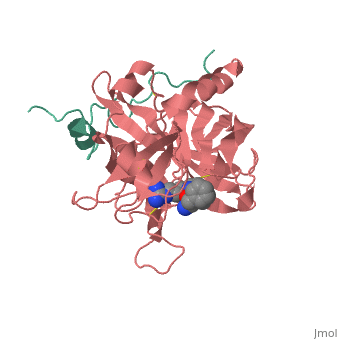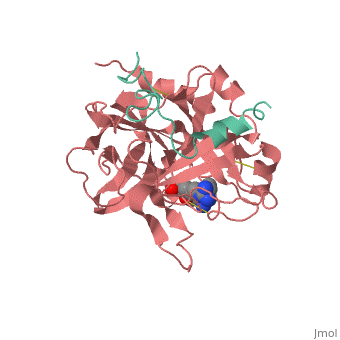
| - | < | + | <StructureSection load='1ppb' size='450' side='right' scene='' caption='α-thrombin large and small subunit complex with phe-pro-arg chloromethylketone (PDB code [[1ppb]])'> |
| - | caption= | + | ===Samer Kawak and Seth Bawel=== |
'''Serine proteases''', or '''proteinases''', so called due to the presence of a serine residue in the active site, are a class of enzymes that catalyse the hydrolysis of peptide bonds in proteins. | '''Serine proteases''', or '''proteinases''', so called due to the presence of a serine residue in the active site, are a class of enzymes that catalyse the hydrolysis of peptide bonds in proteins. | ||
| Line 8: | Line 8: | ||
The B chain consists of <scene name='Serine_Protease/Domains/1'>two domains</scene>. As is true for all of the "trypsin-like" serine proteases, each of the two thrombin domains consists mainly of a 6-stranded, antiparallel beta barrel. The specificity pocket (here filled with the Lys sidechain of the PPACK inhibitor) is in one side of the throat of the domain 2beta barrel, and the activation site is close next to it. | The B chain consists of <scene name='Serine_Protease/Domains/1'>two domains</scene>. As is true for all of the "trypsin-like" serine proteases, each of the two thrombin domains consists mainly of a 6-stranded, antiparallel beta barrel. The specificity pocket (here filled with the Lys sidechain of the PPACK inhibitor) is in one side of the throat of the domain 2beta barrel, and the activation site is close next to it. | ||
| - | |||
| - | <applet load="2ptc" size="350" color="white" frame="true" align="right" spinBox="true" | ||
| - | caption="Trypsin BPT1 complex" /> | ||
==Trypsin-BPTI complex== | ==Trypsin-BPTI complex== | ||
| Line 16: | Line 13: | ||
==succinyl-AAPK-trypsin acyl-enzyme== | ==succinyl-AAPK-trypsin acyl-enzyme== | ||
| + | |||
The atomic resolution structures of trypsin acyl-enzymes like suc-AAPK-trypsin, a tetrahedral intermediate analong, and the Michaelis complex make up the recreation of the catalytic cycle of the serine protease. In a 2006 study by Koshland et al., a model of suc-AAPK-trypsin was studied at 1.28 Å resolution, which exhibited an acyl-enzyme occupancy of 70% with the residual 30% of trypsin extant in free enzyme form. [1] | The atomic resolution structures of trypsin acyl-enzymes like suc-AAPK-trypsin, a tetrahedral intermediate analong, and the Michaelis complex make up the recreation of the catalytic cycle of the serine protease. In a 2006 study by Koshland et al., a model of suc-AAPK-trypsin was studied at 1.28 Å resolution, which exhibited an acyl-enzyme occupancy of 70% with the residual 30% of trypsin extant in free enzyme form. [1] | ||
In suc-AAPK-trypsin, the substrate binds near Ser-195 and His-57. His-57 displays two conformations with equal occupancy. The first conformation positions His-57 near Ser-195 and within hydrogen bonding distance of a water molecule. The water molecule is positioned between His-57 and the acyl carbonyl carbon. In this position, the water molecule is poised for hydrolytic attack. His-57 deprotonates water, turning the water molecule into a better nucleophile. The water molecule is already positioned to directly attack the acyl carbonyl carbon. The <scene name='Ann_Taylor_115/Second_conformation/1'>second conformation</scene> rotates His-57 around the alpha and beta carbons, away from Ser-195 and out of hydrogen bonding distance with the water. This conformation shift takes His-57 out of play. The conformation flexibility of His-57 may be involved in substrate binding or release. [1] | In suc-AAPK-trypsin, the substrate binds near Ser-195 and His-57. His-57 displays two conformations with equal occupancy. The first conformation positions His-57 near Ser-195 and within hydrogen bonding distance of a water molecule. The water molecule is positioned between His-57 and the acyl carbonyl carbon. In this position, the water molecule is poised for hydrolytic attack. His-57 deprotonates water, turning the water molecule into a better nucleophile. The water molecule is already positioned to directly attack the acyl carbonyl carbon. The <scene name='Ann_Taylor_115/Second_conformation/1'>second conformation</scene> rotates His-57 around the alpha and beta carbons, away from Ser-195 and out of hydrogen bonding distance with the water. This conformation shift takes His-57 out of play. The conformation flexibility of His-57 may be involved in substrate binding or release. [1] | ||
| - | |||
| - | <applet load="2agg" size="350" color="white" frame="true" align="right" spinBox="true" | ||
| - | caption="suc-AAPK-trypsin acyl enzyme" /> | ||
{{Clear}} | {{Clear}} | ||
| Line 27: | Line 22: | ||
* Content for this page has been included and adapted with permission from Jane S. and David C. Richardson's http://kinemage.biochem.duke.edu/ | * Content for this page has been included and adapted with permission from Jane S. and David C. Richardson's http://kinemage.biochem.duke.edu/ | ||
| - | + | </StructureSection> | |
==Reference== | ==Reference== | ||
[1] Radisky, E.S., Lee, J.M., Lu C.K., and Koshland, D.E., Jr. ''Proc. Natl. Acad. Sci.'' USA '''2006''', 6835-6840. | [1] Radisky, E.S., Lee, J.M., Lu C.K., and Koshland, D.E., Jr. ''Proc. Natl. Acad. Sci.'' USA '''2006''', 6835-6840. | ||
Current revision
| |||||||||||
Reference
[1] Radisky, E.S., Lee, J.M., Lu C.K., and Koshland, D.E., Jr. Proc. Natl. Acad. Sci. USA 2006, 6835-6840.