Sandbox reserved 330
From Proteopedia
| (38 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
{{STRUCTURE_1ne8| PDB=1ne8 | SCENE= }} | {{STRUCTURE_1ne8| PDB=1ne8 | SCENE= }} | ||
| + | ==Overview of YdcE== | ||
| + | The Bacillus subtilis YdcE gene encodes an endoribonuclease called EndoA, which is a member of the MazF/PemK family of bacterial toxin and the protein encoded by the YdcD gene is an inhibitor of its activity<ref name="Pellegrini"/>. EndoA cleaves at the UAC sequence, which is predicted to be a single stranded conformation, and has an overlapping cleavage site specificity with the MazF E.coli homologues. EndoA activity results in cleavage products with a 3’phosphate and 5’OH group, which is typical of degradative RNAses that functions in the absence of divalent cations <ref name="Pellegrini"> Pellegrini, O., Mathy, N., Gogos.A., Shapiro, L., Condon, C. The Bacillus subtilis YdcDE operon encodes an endoribonuclease of the MazF/PemK family and its inhibitor. Molecular Microbiology.2005. June;56(5):1139-1148</ref>. | ||
| - | == Background Information== | ||
| + | __TOC__ | ||
| - | - | + | == Background on Toxin and Anti-toxin complex== |
| - | + | Addiction modules, consisting of a toxin and antitoxin pair, are controlled by operons which, are autoregulated at the transcriptional level. Bacteria rely on addiction modules to maintain plasmids within populations, and cells that do not inherit the plasmid encoded operon will not produce antitoxin and will be inhibited by the toxin via post segregational killing<ref name="Pellegrini"/>. Once this operon is expressed, the bacterial strain is addicted to the antitoxin for survival. It is known that genomes of most bacteria have a toxin-antitoxin loci, which have been shown to be induced by stressful conditions<ref name="Pellegrini"/>. So thus, these modules play an important role in plasmid partitioning and cellular response to stress, where the maintenance of these modules prevents the lethal effect of toxin on cells. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | - | + | |
| - | + | ||
| - | + | ||
| + | Previous studies of toxin families include MazF, ChpAK, and PemK, which all code for endoribonuclease that activates cellular mRNAs by cleaving them at specific sites. Recently, there is a Bacillus subtilis gene product discovered, EndoA, that is a member of RNAses, which is the gene product of the YdcE gene. This EndoA has similar cleavage pattern specificity as MazF and PemK, with cleavage products of a 3’phosphate and 5’OH group<ref name="Pellegrini"/>. Further study revealed that a coexpression of an upstream gene, YdcD reverses the effects of the YdcE toxin, and thus, this is the first toxin-antitoxin system identified for Bacillus subtilis. However, further research is necessary to determine the functionality of the toxin-antitoxin complex. | ||
| - | - operons encode stable toxin and their antidote | ||
| - | - opersons play an important role in plasmid partitioning and cellular response to stress. | ||
| - | - Family of toxins (MazF/ChpAK/PemK) encodes endoribonuclease that activates cellular mRNAs by cleaving them at specific sites | ||
| - | - This study, there is a B.subtilis gene encode a member of RNases that they call Endo A. | ||
| - | - A coexpression of an upstream gene (YdcD) reverses toxin. | ||
| - | - YdcD inhibits EndoA activity. | ||
| - | - endoA has similar cleavage patterns specificity to MazF/PemK, with cleavage produces with a 3’P and 5’OH group. | ||
| - | - This is the first example of an antitoxin-toxin system of B. subtilis. | ||
| - | |||
| - | - bacteria rely on addiction modules to maintain plasmids within populations. | ||
| - | - These modules consist of operon where one cistron encodes a stable toxin and is preceded by an encoding unstable antitoxin. | ||
| - | - Cells that do not inherit the plasmid encoded operon will not produce antitoxin and inhibited by the toxin via post segregational killing. | ||
| - | - So once this operon is expressed, the bacterial strain is addicted to the antitoxin for survival. | ||
| - | - It is known that genomes of most bacteria has a toxin-antitoxin TA loci. | ||
| - | - These loci have been shown to be induced by stressful conditions. | ||
| - | - It has been shown that antitoxin can also inhibit its own expression by binding to its own promoter region with complexed toxin as corepressor. | ||
| - | - | ||
==Structure== | ==Structure== | ||
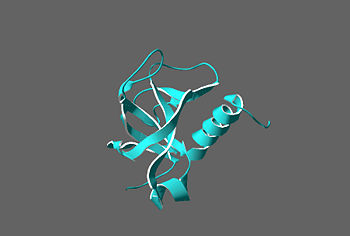
| - | <scene name='Sandbox_reserved_330/ | + | [[Image:Secondary structures.jpg| thumb | left | 350px |Secondary structures of YdcE]] |
| + | Crystallization of the YdcE gene product revealed a crystal with space group of P6522, where a= 56.63, b=56.63, and c=138.257. The final structure model of the YdcE protein was determined to be 2.1 angstroms with an R-factor of 15.9%. The YdcE protein consists of 117 amino acids and is approximately 14 kDa<ref name="Gogos">PMID:14517982</ref>. It is a compact single domain alpha/beta protein, with 3 α helices and 7 β strands. Five out of seven beta strands, β1, β2, β3, β6, and β7 forms an antiparallel sheet. While two of the remaining strands, β4, β5, and C terminus (containing Asp115) of the β3 strand forms a smaller sheet<ref name="Gogos"/>. | ||
| + | |||
| + | |||
| + | The structure itself is a dimer interface between monomers that is related by a two fold axis, and it exists as a dimer in solution as well. The dimer is a convex surface with a flat surface that includes 3 α helix that has C-terminal tails protruding. The convex surface is an extensive hydrophobic surface between the two monomers, and include <scene name='Sandbox_reserved_330/Hydrophobic_residues/1'>Ile 30, Ile 43, Ile 111, Leu 107, Ile 80 and Ile 114.</scene> Each monomer has a β6 strand that is paired with each other through hydrogen bonds between the amide of the Thr82 and the carbonyl oxygen of Ile 80. | ||
| + | |||
| + | |||
| + | On the convex side of the dimer, hydrogen bonds exist between amides of Ser 19, to the side chain of Asp 84, along with salt bridges between Glu 20 and Arg 87<ref name="Gogos"/>. Between these salt bridges, the Arg 81 of each monomer are buried in the dimer interface and is stabilized by water-mediated hydrogen bonds. Other dimer interactions of the YdcE protein include a hydrogen bond between carbonyl oxygen of Ser 110 and the amide of Asn 32, and between the carbonyl oxygen of Ala 112 and NE of Arg5<ref name="Gogos"/>. | ||
| + | |||
| + | |||
| + | [[Image:6_charged_AA.jpg | thumb | right | 400px |6 charged amino acids contributing to electronegative surface potential]] | ||
| + | The YdcE protein has similar structures to other proteins, such as Kid from E.coli in plasmid R1, and CcdB from E.coli in plasmid F. These similarities include a five stranded antiparallel sheet and a smaller three stranded β-sheet with a C-terminal α helix<ref name="Gogos"/>. YdcE shares 27% sequence similarity with Kid and 7% with CcdB. However, the electronegative surface potential of YdcE is more negative than Kid and CcdB, with a pI of 4.7<ref name="Almrud"> Almrud, J.J., Kern, A.D., Wang, S.C., Czerwinski, R.M., Johnson, W.H., Murzin, A.G., Hackert, M.L., Whitman, C.P. The crystal structure of YdcE, a 4-oxalocrotonate tautomerase homologue from Escherichia coli., confirms the structural basis for oligomer diversity. Biochemistry.2002. August;41(40):12010-12024</ref>. This is largely due to having six charged amino acids; Asp 96, Asp 97, Glu 98, Glu 105, Asp 101, and Asp 104<ref name="Gogos"/>. | ||
| + | |||
| + | |||
| + | ==Active Site== | ||
| - | + | Complexes of YdcE reveal that the two active sites of the enzyme are located peripherally at the dimer interface, and are shown to be composed of residues contributed from both monomers of the dimer. The two active sites of the native YdcE protein structure have a few differences to those when complexed with other proteins, however, the largest difference is in the repositioning of the aromatic ring of the Phe8, which is rotated approximately by 32° in the complex structure relative to the native YdcE protein<ref name="Almrud"/>. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | The active site of the YdcE protein is composed of residues from both monomers, with key active site residues consisting of Pro1, Arg 11, Arg 38, Phe50. Dimerization of the two monomers include Pro1, which is presumed to be the catalytic base and is from one subunit, while Phe8, Arg 10, Trp 51, and Tyr72 are from the other monomer<ref name="Almrud"/>. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | - the protein surface and C-terminus of YdcE protein is involved in toxin interaction with it’s target. | ||
| - | - The C-terminus of these homologs vary, so variability may reflect substrate specificity within the protein family. | ||
| - | <ref>PMID:14517982</ref> | ||
| - | ==Function== | ||
<references/> | <references/> | ||
Current revision
| |||||||||
| 1ne8, resolution 2.10Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||||
| Gene: | ydcE (Bacillus subtilis) | ||||||||
| |||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum, TOPSAN | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Overview of YdcE
The Bacillus subtilis YdcE gene encodes an endoribonuclease called EndoA, which is a member of the MazF/PemK family of bacterial toxin and the protein encoded by the YdcD gene is an inhibitor of its activity[1]. EndoA cleaves at the UAC sequence, which is predicted to be a single stranded conformation, and has an overlapping cleavage site specificity with the MazF E.coli homologues. EndoA activity results in cleavage products with a 3’phosphate and 5’OH group, which is typical of degradative RNAses that functions in the absence of divalent cations [1].
Contents |
Background on Toxin and Anti-toxin complex
Addiction modules, consisting of a toxin and antitoxin pair, are controlled by operons which, are autoregulated at the transcriptional level. Bacteria rely on addiction modules to maintain plasmids within populations, and cells that do not inherit the plasmid encoded operon will not produce antitoxin and will be inhibited by the toxin via post segregational killing[1]. Once this operon is expressed, the bacterial strain is addicted to the antitoxin for survival. It is known that genomes of most bacteria have a toxin-antitoxin loci, which have been shown to be induced by stressful conditions[1]. So thus, these modules play an important role in plasmid partitioning and cellular response to stress, where the maintenance of these modules prevents the lethal effect of toxin on cells.
Previous studies of toxin families include MazF, ChpAK, and PemK, which all code for endoribonuclease that activates cellular mRNAs by cleaving them at specific sites. Recently, there is a Bacillus subtilis gene product discovered, EndoA, that is a member of RNAses, which is the gene product of the YdcE gene. This EndoA has similar cleavage pattern specificity as MazF and PemK, with cleavage products of a 3’phosphate and 5’OH group[1]. Further study revealed that a coexpression of an upstream gene, YdcD reverses the effects of the YdcE toxin, and thus, this is the first toxin-antitoxin system identified for Bacillus subtilis. However, further research is necessary to determine the functionality of the toxin-antitoxin complex.
Structure
Crystallization of the YdcE gene product revealed a crystal with space group of P6522, where a= 56.63, b=56.63, and c=138.257. The final structure model of the YdcE protein was determined to be 2.1 angstroms with an R-factor of 15.9%. The YdcE protein consists of 117 amino acids and is approximately 14 kDa[2]. It is a compact single domain alpha/beta protein, with 3 α helices and 7 β strands. Five out of seven beta strands, β1, β2, β3, β6, and β7 forms an antiparallel sheet. While two of the remaining strands, β4, β5, and C terminus (containing Asp115) of the β3 strand forms a smaller sheet[2].
The structure itself is a dimer interface between monomers that is related by a two fold axis, and it exists as a dimer in solution as well. The dimer is a convex surface with a flat surface that includes 3 α helix that has C-terminal tails protruding. The convex surface is an extensive hydrophobic surface between the two monomers, and include Each monomer has a β6 strand that is paired with each other through hydrogen bonds between the amide of the Thr82 and the carbonyl oxygen of Ile 80.
On the convex side of the dimer, hydrogen bonds exist between amides of Ser 19, to the side chain of Asp 84, along with salt bridges between Glu 20 and Arg 87[2]. Between these salt bridges, the Arg 81 of each monomer are buried in the dimer interface and is stabilized by water-mediated hydrogen bonds. Other dimer interactions of the YdcE protein include a hydrogen bond between carbonyl oxygen of Ser 110 and the amide of Asn 32, and between the carbonyl oxygen of Ala 112 and NE of Arg5[2].
The YdcE protein has similar structures to other proteins, such as Kid from E.coli in plasmid R1, and CcdB from E.coli in plasmid F. These similarities include a five stranded antiparallel sheet and a smaller three stranded β-sheet with a C-terminal α helix[2]. YdcE shares 27% sequence similarity with Kid and 7% with CcdB. However, the electronegative surface potential of YdcE is more negative than Kid and CcdB, with a pI of 4.7[3]. This is largely due to having six charged amino acids; Asp 96, Asp 97, Glu 98, Glu 105, Asp 101, and Asp 104[2].
Active Site
Complexes of YdcE reveal that the two active sites of the enzyme are located peripherally at the dimer interface, and are shown to be composed of residues contributed from both monomers of the dimer. The two active sites of the native YdcE protein structure have a few differences to those when complexed with other proteins, however, the largest difference is in the repositioning of the aromatic ring of the Phe8, which is rotated approximately by 32° in the complex structure relative to the native YdcE protein[3].
The active site of the YdcE protein is composed of residues from both monomers, with key active site residues consisting of Pro1, Arg 11, Arg 38, Phe50. Dimerization of the two monomers include Pro1, which is presumed to be the catalytic base and is from one subunit, while Phe8, Arg 10, Trp 51, and Tyr72 are from the other monomer[3].
- ↑ 1.0 1.1 1.2 1.3 1.4 Pellegrini, O., Mathy, N., Gogos.A., Shapiro, L., Condon, C. The Bacillus subtilis YdcDE operon encodes an endoribonuclease of the MazF/PemK family and its inhibitor. Molecular Microbiology.2005. June;56(5):1139-1148
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Gogos A, Mu H, Bahna F, Gomez CA, Shapiro L. Crystal structure of YdcE protein from Bacillus subtilis. Proteins. 2003 Nov 1;53(2):320-2. PMID:14517982 doi:http://dx.doi.org/10.1002/prot.10457
- ↑ 3.0 3.1 3.2 Almrud, J.J., Kern, A.D., Wang, S.C., Czerwinski, R.M., Johnson, W.H., Murzin, A.G., Hackert, M.L., Whitman, C.P. The crystal structure of YdcE, a 4-oxalocrotonate tautomerase homologue from Escherichia coli., confirms the structural basis for oligomer diversity. Biochemistry.2002. August;41(40):12010-12024