Hen Egg-White (HEW) Lysozyme
From Proteopedia
| (213 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | __TOC__ | |
| - | Lysozyme | + | [[Image:1hewB.gif|frame|Hen egg-white lysozyme, 1HEW (scroll down to the [[#Structure_of_Lysozyme|structure section]] for interactive figure)]] |
| - | + | '''Lysozyme''' - also known as muramidase - is a powerful enzyme found in abundance in tears, saliva, and human milk. In humans, it is encoded in the ''LYZ'' gene. Since it is a small, easily available, and highly stable protein, it has been subject to extensive research regarding its function and structure. | |
| - | + | ||
| - | + | ==Introduction== | |
| - | < | + | Lysozyme acts as a non-specific defense against bacteria and fungi. It is a component of the innate immune system, and is an important part of an infant's diet to ward off diarrhea. It is an enzyme known for its ability to degrade the polysaccharide architecture of many kinds of cell walls, normally for the purpose of protection against bacterial infection<ref>PMID 28934357</ref>. The structure of hen egg white (HEW) lysozyme, the focus of this article, is shown on the right. The antibacterial activity of hen egg white was first described by Laschtschenko in 1909<ref>Laschtschenko,P. (1909) |
| + | Über die keimtötende und entwicklungshemmende Wirkung von Hühnereiweiss. Z. Hyg. Infektionskrankh.,64,419-427. | ||
| + | </ref>. It was characterized and named “lysozyme” by Alexander Fleming, the same person credited for the discovery of penicillin<ref>Fleming, A. (1922) On a remarkable bacteriolytic element found in tissues and secretions. Proc.Roy.Soc.(London),93,306-317.</ref>. Discovery of the enzymatic activity was by accident; during the unrelated experiment, nasal drippings were inadvertently introduced to a petri dish containing a bacterial culture, which culture consequently exhibited the results of an as yet unknown enzymatic reaction. The observation of this unknown reaction led to further research on the components of this reaction as well as to the corresponding identification of the newfound "lysozyme." In 1965, David C. Phillips and coworkers determined the three-dimensional structure of lysozyme at 2 Å resolution <ref>PMID 5891407</ref>. Phillips' work was especially groundbreaking since Phillips had managed to successfully elucidate the structure of an enzyme via X-ray crystallography - a feat that had never before been accomplished<ref>Bugg, T. 1997. An Introduction to Enzyme and Coenzyme Chemistry. Blackwell Science Ltd., Oxford </ref><ref name="earliest">[http://www.umass.edu/microbio/rasmol/1st_xtls.htm Earliest Solutions for Macromolecular Crystal Structures].</ref>. Phillips' research also led to a structure-based hypothesis of its mechanism of action | ||
| + | <ref name=Phillips1967>PMID 4382801</ref>.<br /> | ||
| + | ==Function== | ||
| + | The primary function of lysozyme is to hydrolyze the bonds between the sugar molecules in the peptidoglycan layer of bacterial cell walls. Peptidoglycan is a rigid structure that provides support and protection to bacterial cells. By cleaving these bonds, lysozyme weakens the bacterial cell wall, leading to the rupture and death of the bacterium. | ||
| - | + | Lysozyme is particularly effective against Gram-positive bacteria, which have a relatively thick peptidoglycan layer. Gram-negative bacteria, on the other hand, have an outer membrane that provides additional protection, making them less susceptible to lysozyme. | |
| + | In addition to its antimicrobial activity, lysozyme also plays a role in other physiological processes. It contributes to the maintenance of healthy epithelial tissues, such as those lining the respiratory and gastrointestinal tracts. Lysozyme helps prevent bacterial overgrowth and infection in these areas by inhibiting the growth of certain bacteria. | ||
| + | Lysozyme is widely distributed in nature and can be found in various organisms, including humans, animals, and plants. It is particularly abundant in the secretions of the lacrimal glands (tears) and salivary glands, where it helps protect the eyes and oral cavity from bacterial infections. | ||
| + | Due to its antimicrobial properties, lysozyme has been used in various applications, such as a natural food preservative and an additive in certain personal care products. It has also been studied for its potential therapeutic applications, including in the development of antimicrobial agents and the treatment of certain infections. | ||
| - | + | *'''Tail-associated lysozyme''' is expressed in bacteriophage T4 plays a role in digesting the peptidoglycan layering in lysis from within<ref>PMID: 3157805</ref>. | |
| - | == | + | ==Chemical activity== |
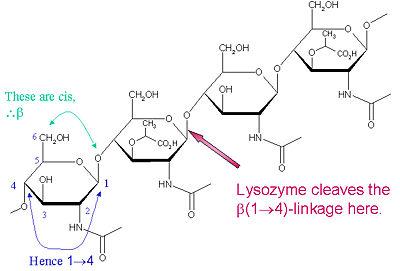
| + | [[Image:nag-nam2.jpg|left|400px|Lysozyme Cleavage Site<ref>Image from: http://www.vuw.ac.nz/staff/paul_teesdale-spittle/essentials/chapter-6/proteins/lysozyme.htm</ref>]] | ||
| - | + | The particular substrate of preference for this cleavage type is a (NAG-NAM)₃ hexasaccharide, within which substrate occurs the | |
| - | + | cleaving target glycosidic bond, NAM₄-β-O-NAG₅. The individual hexasaccharide binding units are designated A-F, with NAM₄-β-O-NAG₅ glycosidic bond cleavage preference corresponding to a D-E unit glycosidic bond cl | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Lysozyme is known for damaging bacterial cell walls by catalyzing the hydrolysis of 1,4-beta-linkages between N-acetylmuramic acid (NAM) and N-acetyl-D-glucosamine (NAG) residues in peptidoglycan, and between N-acetyl-D-glucosamine residues in chitodextrins. In this way, lysozyme is efficient in lysing the cell walls of both bacteria and fungi. The location of cleavage for lysozyme on this architectural theme is the β(1-4) glycosidic linkage connecting the C1 carbon of NAM to the C4 carbon of NAG. | |
| - | + | The particular substrate of preference for this cleavage type is a (NAG-NAM)₃ hexasaccharide, within which substrate occurs the | |
| - | + | cleaving target glycosidic bond, NAM₄-β-O-NAG₅. The individual hexasaccharide binding units are designated A-F, with NAM₄-β-O-NAG₅ glycosidic bond cleavage preference corresponding to a D-E unit glycosidic bond cleavage preference. Depending on the organism from which lysozyme is obtained, hydrolysis of the glycosidic bond proceeds with retention of configuration at the anomeric carbon (hen egg white) or with inversion (goose, phage T4). | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Lysozyme efficiently acts on long (NAG-NAM) or (NAG) polymers. As the chain length gets smaller than six monomers, the catalytic rates drop substantially; in fact, trisaccharides act as competitive inhibitors by binding to the active site in a non-productive register. | |
| - | + | <StructureSection load='' size='400' side='right' scene='37/376372/Overall/3' caption='Hen egg white lysozyme (PDB code [[1hew]])'> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | == Structure of Lysozyme == |
| - | + | <jmol><jmolLink><script>script /scripts/37/376372/Overall/3.spt; hide water; set zshade off; spin off;</script><text>Lysozyme</text></jmolLink> </jmol> (shown here: PDB code 1HEW) is a small single-[[chain]] protein containing 129 [[amino acids]]. It folds into a compact structure with an active site cleft that binds to certain carbohydrates (<jmol><jmolLink><script> select ligand; selectionHalos ON; delay 0.5;selectionHalos OFF;</script><text>☼</text></jmolLink> </jmol>). Starting at the blue end, the N terminus(<jmol><jmolLink><script> select 1.CA; selectionHalos ON; delay 0.5;selectionHalos OFF;</script><text>☼</text></jmolLink> </jmol>), one can trace the entire chain (<jmol><jmolLink><script>select *.ca and 1; selectionhalos on; | |
| - | + | delay 0.5; | |
| - | + | select *.ca and 1-10; delay 0.3; | |
| - | [[ | + | select *.ca and 5-15; delay 0.2; |
| - | + | select *.ca and 11-20; delay 0.1; | |
| - | + | select *.ca and 15-25; delay 0.1; | |
| - | + | select *.ca and 21-30; delay 0.1; | |
| - | + | select *.ca and 25-35; delay 0.1; | |
| - | + | select *.ca and 31-40; delay 0.1; | |
| - | + | select *.ca and 35-45; delay 0.1; | |
| - | + | select *.ca and 41-50; delay 0.1; | |
| - | + | select *.ca and 45-55; delay 0.1; | |
| - | + | select *.ca and 51-60; delay 0.1; | |
| - | + | select *.ca and 55-65; delay 0.1; | |
| + | select *.ca and 61-70; delay 0.1; | ||
| + | select *.ca and 65-75; delay 0.1; | ||
| + | select *.ca and 71-80; delay 0.1; | ||
| + | select *.ca and 75-85; delay 0.1; | ||
| + | select *.ca and 81-90; delay 0.1; | ||
| + | select *.ca and 85-95; delay 0.1; | ||
| + | select *.ca and 91-100; delay 0.1; | ||
| + | select *.ca and 101-110; delay 0.1; | ||
| + | select *.ca and 111-122; delay 0.1; | ||
| + | select *.ca and 122-127; delay 0.2; | ||
| + | select *.ca and 124-129; delay 0.3; | ||
| + | select *.ca and 129; delay 0.5; | ||
| + | selectionhalos off; </script><text>☼</text></jmolLink> </jmol>) through to the C terminus (<jmol><jmolLink><script> select 129.CA; selectionHalos ON; delay 0.5;selectionHalos OFF;</script><text>☼</text></jmolLink> </jmol>), showing the folding pattern of the protein. There are four disulfide bridges (between Cys 6 and Cys 127, between Cys 30 and Cys 115, between Cys 64 and Cys 80 and between Cys 76 and Cys 94 <jmol><jmolLink><script> select BONDS ({0:3}); wireframe 0.6; delay 0.5; wireframe 0.1</script><text>☼</text></jmolLink> </jmol>) that crosslink the polypeptide, stabilizing the native conformation. Lysozyme's secondary structure comprises four alpha (and one 3-10) helices (<jmol><jmolLink><script> select helix and *.CA; selectionHalos ON; delay 0.5;selectionHalos OFF;</script><text>☼</text></jmolLink> </jmol>)and five beta strands (<jmol><jmolLink><script> select sheet and *.CA; selectionHalos ON; delay 0.5;selectionHalos OFF;</script><text>☼</text></jmolLink> </jmol>). Linking these, a number of beta turns and a large number of random coils make up the remainder of the polypeptide backbone. The [[secondary structure]] is easier to see in a <scene name='37/376372/Secondary_structure/7'>ribbon view</scene>. | ||
| - | = | + | However, this type of [[backbone representation]] ([https://stories.duke.edu/sciences-mother-of-ribbon-diagrams-celebrates-50-years-at-duke Richardson diagram]) was not invented until about a quarter century after this first enzyme structure was solved<ref name="jr1">PMID:10932243</ref>, so the figures in the original report look more like the previous scene. |
| - | + | === Surface properties === | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | The shape and the properties of the <jmol><jmolLink><script> script /scripts/37/376372/Surface/4.spt; background white; hide water or ligand</script><text>surface of lysozyme</text></jmolLink> </jmol> determines what it binds to. When the protein folds into its native structure, the hydrophobicity and the charge of the side chains are a driving force. <jmol> | |
| - | + | <jmolLink> | |
| + | <script>select backbone or water; spacefill off; hide ligand or (sidechain and not (met, cys, ile, leu, val, phe, tyr, trp))</script> | ||
| + | <text>Hydrophobic residues</text> | ||
| + | </jmolLink> | ||
| + | </jmol> (shown here: Met, Cys, Leu, Val, Ile, Phe, Tyr, and Trp) occur mostly on the interior of the enzyme while <jmol> | ||
| + | <jmolLink> | ||
| + | <script>select backbone or water; spacefill off; hide ligand or (sidechain and (met, cys, ile, leu, val, phe, tyr, trp))</script> | ||
| + | <text>hydrophilic residues and proline</text> | ||
| + | </jmolLink> | ||
| + | </jmol> tend to be on the surface of the protein. In the crystal structure as well as in aqueous solution, <jmol> | ||
| + | <jmolLink> | ||
| + | <script>select backbone; spacefill on; select water; spacefill 1.0; display all</script> | ||
| + | <text>water</text> | ||
| + | </jmolLink></jmol> binds to the surface of the enzyme, as does the substrate and the product of the catalyzed hydrolysis reaction. To explore these aspects of protein structure and function, switch different parts of the structure on and off and change the coloring scheme (side chain polarity or charge) using the buttons below. | ||
| - | + | <jmol> | |
| + | <jmolRadioGroup> | ||
| + | <item> | ||
| + | <script>hide (hidden and not sidechain) or (sidechain and not (met, cys, ile, leu, val, phe, tyr, trp))</script> | ||
| + | <text>hydrophobic side chains</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>hide (hidden and not sidechain) or (sidechain and (met, cys, ile, leu, val, phe, tyr, trp))</script> | ||
| + | <text>hydrophilic side chains</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>display (displayed and not sidechain) or sidechain</script> | ||
| + | <text>all side chains</text> | ||
| + | <checked>true</checked> | ||
| + | </item> | ||
| + | </jmolRadioGroup> | ||
| + | </jmol> | ||
| - | === | + | <jmol> |
| + | <jmolRadioGroup> | ||
| + | <item> | ||
| + | <script>select sidechain; color magenta; select sidechain and (cys, met, ile, leu, val, phe, tyr, trp); color gray; set echo ID bla 80% 0%; echo "hydrophobic"; color echo gray; frank off; set echo ID bla2 0% 0%; echo "hydrophilic"; color echo magenta </script> | ||
| + | <text>colored by hydrophobiticity</text> | ||
| + | <checked>true</checked> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>select sidechain; color white; select sidechain and (asp, glu); color red; select sidechain and (lys, arg, his); color blue; set echo ID bla 80% 0%; echo "positive"; color echo blue; frank off; set echo ID bla2 0% 0%; echo "negative"; color echo red</script> | ||
| + | <text>colored by charge</text> | ||
| + | <checked>false</checked> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script> select sidechain; define ~consurf_to_do selected; define ~consurf_to_color selected; useFullScript = true; consurf_initial_scene = false; script /wiki/ConSurf/he/1hew_consurf.spt; set echo ID bla 80% 0%; echo "conserved"; color echo palevioletred; frank off; set echo ID bla2 0% 0%; echo "variable"; color echo darkturquoise</script> | ||
| + | <text>colored by conservation</text> | ||
| + | <checked>false</checked> | ||
| + | </item> | ||
| + | </jmolRadioGroup> | ||
| + | </jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>if ({*.O and backbone and visible}.size>0) {select backbone; spacefill off} else {select backbone; spacefill on} </script><text>[± backbone spacefill]</text></jmolLink> </jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>if ({ligand and hidden}.size>0) {display displayed or ligand} else {hide ligand or hidden} </script><text>[± ligand]</text></jmolLink> </jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>if ({water and visible}.size>0) {select water; spacefill off} else {select water; spacefill 1.0} </script><text>[± water]</text></jmolLink> </jmol> | ||
| + | |||
| + | ===Hydrogen Bonding=== | ||
| + | |||
| + | In all proteins <jmol><jmolLink> | ||
| + | <script>set picking center; display all; script /scripts/37/376372/Hbonds/6.spt </script> | ||
| + | <text>hydrogen bonds</text> | ||
| + | </jmolLink></jmol> are essential for stability and function. Main chain hydrogen bonds are found within <jmol><jmolLink> | ||
| + | <script>script /scripts/37/376372/Hbonds/1.spt; select helix and backbone; hbonds 0.2; delay 1.2; hbonds 0.04; </script> | ||
| + | <text>helices</text> | ||
| + | </jmolLink></jmol>, in <jmol><jmolLink> | ||
| + | <script>script /scripts/37/376372/Hbonds/5.spt; select 74,77; hbonds 0.2; delay 1.2; hbonds 0.04; </script> | ||
| + | <text>beta turns</text> | ||
| + | </jmolLink></jmol> and <jmol><jmolLink> | ||
| + | <script>script /scripts/37/376372/Hbonds/2.spt; select sheets and backbone; hbonds 0.2; delay 1.2; hbonds 0.04; </script> | ||
| + | <text>between strands</text> | ||
| + | </jmolLink></jmol>, connecting them to form sheets. <jmol> | ||
| + | <jmolLink> | ||
| + | <script>hide none; script /scripts/37/376372/Hbonds/4.spt </script> | ||
| + | <text>Ordered waters</text> | ||
| + | </jmolLink> | ||
| + | </jmol> found in the crystal structure are bound to the protein surface by one or more hydrogen bonds. The specificity of the enzyme active site is partially determined by the hydrogen bonds <jmol> | ||
| + | <jmolLink> | ||
| + | <script>script /scripts/37/376372/Hbonds/3.spt; select ligand and not 201:A.O1; set bondMode OR; hbonds 0.2; delay 1.2; hbonds 0.04; </script> | ||
| + | <text>between protein and ligand</text> | ||
| + | </jmolLink> | ||
| + | </jmol>. | ||
| + | |||
| + | This is the only scene that shows all atoms in the structure. Two tricks are used to improve the clarity of the scene. First, the atoms in the back (with respect to the current view) are faded. The same trick is used in this [https://cdn.rcsb.org/pdb101/geis/images/1000w/geis-0512-lysozyme.png painting] of the structure by Irving Geis. If you rotate the view to bring these atoms to the front, they will become bright and others will be faded. Second, the atoms in the very front are removed from view (slab). However, if you rotate the molecule they will come into view. | ||
| + | |||
| + | <jmol> | ||
| + | <jmolButton> | ||
| + | <script>if (zshade) {set zshade off; slab off} else {set zshade on; slab on}</script> | ||
| + | <text>toggle fading and slabbing</text> | ||
| + | </jmolButton></jmol> | ||
| + | |||
| + | <jmol> | ||
| + | <jmolButton> | ||
| + | <script>set picking center</script> | ||
| + | <text>turn centering on</text> | ||
| + | </jmolButton></jmol> <jmol> | ||
| + | <jmolButton> | ||
| + | <script>set picking identify</script> | ||
| + | <text>turn centering off</text> | ||
| + | </jmolButton></jmol> | ||
| + | |||
| + | |||
| + | == Ligand-bound structures == | ||
| + | |||
| + | Ligand-bound structures are valuable to understand specificity and mechanism of enzymes. Ideally, these structures include a substrate complex, a transition state analog complex and a product complex. Because lysozyme has a repetitive polymer as substrate and because of its specific binding properties, there is still no substrate complex structure with a glycosidic bond in the correct register. This has made it difficult to study the mechanism based on structural data (see below). | ||
| + | |||
| + | === Inhibitor complex and hypothetical model of substrate complex === | ||
| + | |||
| + | Lysozyme is inhibited by short-length oligosaccharides which act competitively with the natural substrate. The smaller saccharides will bind to the first three binding sites of the cleft (sites A-C), but will not reach sites D and E, where the enzyme cuts the glycosidic bond. So the competitive inhibitor will stick in the cleft, not allowing the substrate to bind to the enzyme complex.<ref>http://mcdb-webarchive.mcdb.ucsb.edu/sears/biochemistry/tw-enz/lysozyme/HEWL/lysozyme-overview.htm</ref>. When the 2 Å structure of lysozyme was reported in 1965<ref> PMID 5891407</ref>, it was published back to back with a report of several 6 Å structures of inhibitor-bound structures<ref> PMID 5840126</ref>. The refined structure of one of these inhibitor structures at higher resolution was deposited as 1HEW in the protein data bank in 1992, and this is the structure all the previous scenes are based on. The bound inhibitor (NAG)<sub>3</sub> occupies subsites A, B, and C. | ||
| + | |||
| + | In order to deduce the catalytic mechanism of lysozyme, the Phillips groups modeled six sugar residues into the active site. Phillips describes <scene name='37/376372/Hex_model/1'>this hypothetical model</scene> in a 1966 article in Scientific American<ref>Phillips (1966) Scientific American 215, 76-90</ref>, and it is featured as a painting by Irving Geis on the [https://pdb101.rcsb.org/sci-art/geis-archive/gallery/geis-0512-lysozyme front cover]. | ||
| + | <jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>display all</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>hide 204-206</scriptWhenUnchecked> | ||
| + | <checked>true</checked> | ||
| + | <text>modeled extra sugars</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol> | ||
| + | |||
| + | === Covalent intermediate and product complex === | ||
| + | |||
| + | In 2001, Stephen Withers and coworkers published a <scene name='37/376372/Covalent_intermediate/2'>covalent intermediate</scene> (PDB code 1H6M) in which Glu 35 had been mutated to Gln to remove the general acid catalyst and the substrate contained NAG-2-fluoro-glucosyl fluoride (NAG2FGlcF) <ref name=Withers2001>PMID 11518970</ref>. The combination of these two changes slowed down the reaction sufficiently to capture the intermediate. This structure and the biochemical results were strong evidence that lysozyme forms a covalent intermediate (via Asp 52) as one step in the reaction. | ||
| + | |||
| + | When lysozyme breaks down peptidoglycan, the product of the reaction is a shortened peptidoglycan that is broken down further by lysozyme. This makes it difficult to crystallize an enzyme: product complex. Vocadlo and coworkers <ref>Davies, Withers and Vocadlo (2009) The Chitopentaose Complex of a Mutant Hen Egg-White Lysozyme Displays No Distortion of the –1 Sugar Away from a 4C1 Chair Conformation, Australian Journal of Chemistry 62(6) 528-532</ref> determined the structure of a <scene name='37/376372/Product_complex/6'>product complex with five sugar units</scene> (PDB code 2WAR) by cocrystallizing the oligosaccharide with an almost inactive mutant of lysozyme (using the same Glu35Gln mutant as used for the covalent intermediate). To compare with the covalent intermediate structure, use the selections below: | ||
| + | |||
| + | <jmol> | ||
| + | <jmolRadioGroup> | ||
| + | <item> | ||
| + | <script>anim off; delay 1.0; model 1</script> | ||
| + | <text>product</text> | ||
| + | <checked>true</checked> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>anim off; delay 1.0; model 2</script> | ||
| + | <text>intermediate</text> | ||
| + | <checked>false</checked> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>anim fps 1; anim mode loop; anim on;</script> | ||
| + | <text>animate</text> | ||
| + | <checked>false</checked> | ||
| + | </item> | ||
| + | </jmolRadioGroup> | ||
| + | </jmol> | ||
| + | |||
| + | <jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>hide ligand and not ((1134 or G2F) and (*.O1, *.C1, *.C2, *.C3, *.C4, *.C5, *.O5, *.N5, *.O4))</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>hide water</scriptWhenUnchecked> | ||
| + | <checked>false</checked> | ||
| + | <text>just ring</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol> | ||
| + | |||
| + | </StructureSection> | ||
| + | |||
| + | <jmol> | ||
| + | <jmolButton> | ||
| + | <script>moveto 2.0 { 579 257 774 163.39} 120.0 0.0 0.0 {-0.33203837 20.866533 19.55904} 31.371292 {0 0 0} 0 0 0 3.0 0.0 0.0;</script> | ||
| + | <text>view 1</text> | ||
| + | </jmolButton></jmol> <jmol> | ||
| + | <jmolButton> | ||
| + | <script>moveto /* time, axisAngle */ 2.0 { -919 -290 -267 175.7} /* zoom, translation */ 120.0 0.0 0.0 /* center, rotationRadius */ {-0.33203837 20.866533 19.55904} 31.371292 /* navigation center, translation, depth */ {0 0 0} 0 0 0 /* cameraDepth, cameraX, cameraY */ 3.0 0.0 0.0; | ||
| + | </script> | ||
| + | <text>view 2</text> | ||
| + | </jmolButton></jmol> <jmol> | ||
| + | <jmolButton> | ||
| + | <script>moveto /* time, axisAngle */ 2.0 { -430 -10 903 146.05} /* zoom, translation */ 120.0 0.0 0.0 /* center, rotationRadius */ {-0.33203837 20.866533 19.55904} 31.371292 /* navigation center, translation, depth */ {0 0 0} 0 0 0 /* cameraDepth, cameraX, cameraY */ 3.0 0.0 0.0;</script> | ||
| + | <text>view 3</text> | ||
| + | </jmolButton></jmol> <jmol> | ||
| + | <jmolButton> | ||
| + | <script>moveto /* time, axisAngle */ 2.0 { 14 -997 -82 164.64} /* zoom, translation */ 615.28 0.0 0.0 /* center, rotationRadius */ {6.174519999999999 24.04584 25.138840000000002} 40.46755187629986 /* navigation center, translation, depth */ {0 0 0} 0 0 0 /* cameraDepth, cameraX, cameraY */ 3.0 0.0 0.0;</script> | ||
| + | <text>🔎 active site</text> | ||
| + | </jmolButton></jmol> <jmol> | ||
| + | <jmolButton> | ||
| + | <script>moveto /* time, axisAngle */ 2.0 { 311 -943 -118 125.09} /* zoom, translation */ 500.88 16.86 6.57 /* center, rotationRadius */ {5.804955513628509 21.50459857126935 17.894926846396874} 35.7306153511559 /* navigation center, translation, depth */ {0 0 0} 0 0 0 /* cameraDepth, cameraX, cameraY */ 3.0 0.0 0.0;</script> | ||
| + | <text>🔎 active site 2</text> | ||
| + | </jmolButton></jmol> <jmol> | ||
| + | <jmolButton> | ||
| + | <script>script http://proteopedia.org/wiki/images/7/76/Wobble.spt</script> | ||
| + | <text>wobble</text> | ||
| + | </jmolButton></jmol> <jmol> | ||
| + | <jmolButton> | ||
| + | <script>script http://proteopedia.org/wiki/images/b/b9/Bobble2.spt</script> | ||
| + | <text>bobble</text> | ||
| + | </jmolButton></jmol> | ||
| + | |||
| + | |||
| + | ==Mechanism== | ||
| + | |||
| + | Hydrolysis of glycosidic bonds by hen egg white lysozyme proceeds with retention of configuration. In 1953, Koshland <ref>Koshland, D. E. (1953). Biol. Rev. 28, 416–436</ref> suggested that in general, retention of configuration implies a double-displacement mechanism (while inversion of configuration implies single displacement). For decades, two competing mechanistic hypotheses (Phillips<ref name=Phillips1967/>: dissociative mechanism with oxocarbenium intermediate; Koshland: two-step associative mechanism with covalent enzyme complex as intermediate) were considered, with data from 2001<ref name=Withers2001/> tipping the scale toward the existence of a covalent intermediate. The absence of a substrate complex structure certainly contributed to difficulties distinguishing between possible mechanisms, as did the existence of two distinct mechanisms (retention and inversion of configuration) within the same structural family of enzymes (e.g. hen vs goose enzyme). | ||
| + | |||
| + | |||
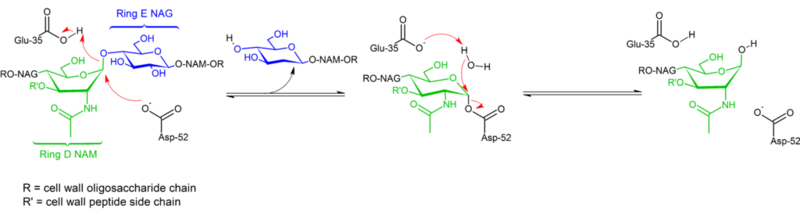
| + | [[Image:Mechanism_of_Lysozyme_action.png|left|800px|Mechanism of Lysozyme]] | ||
| + | {{Clear}} | ||
| + | Lysozyme hydrolyzes a glycoside (hence the familial classification of lysozyme as a glycosylase<ref>https://bio.libretexts.org/Bookshelves/Biochemistry/Book%3A_Biochemistry_Online_(Jakubowski)/07%3A_CATALYSIS/B._Mechanisms_of_Enzyme-Catalyzed_Reactions/B2.__Lysozyme</ref>), which corresponds to the conversion of an acetal to a hemiacetal. The reaction proceeds in two steps as shown in the figure above. In the first step, Asp 52 acts as nucleophile and part of the sugar is the leaving group. In the second step, water acts as nucleophile and Asp 52 acts as leaving group. Both steps invert the configuration at the anomeric carbon, leading to an overall retention of configuration. Glu 35 acts as an acid in the first step (protonating the sugar the the glycosidic bond to make it a better electrophile) and as a base in the second step (deprotonating water to make it a better nucleophile). While the figure shows some of the sugars in a boat conformation to emphasize the inversion of configuration, these are not observed experimentally but are rather found in a chair conformation. | ||
| + | |||
| + | == Applications of Lysozyme == | ||
| + | |||
| + | Since lysozyme has been widely recognized for its antibacterial and antifungal properties, it has a wide variety of uses both in biochemical and pharmaceutical applications. In molecular biology, lysozyme is often used in the alkaline-lysis procedure for extracting and isolating plasmid DNA. It is used extensively in the pharmaceutical field for destroying gram-positive bacteria, and can be used to support already-existing immune defenses to fight bacterial infections. This enzyme is particularly important for preventing bacterial diseases in infants. Because of its antibacterial properties, lysozyme can also be used in the food industry to help prevent spoilage of foods. | ||
| + | |||
| + | ==3D structures of lysozome== | ||
| + | [[Lysozyme 3D structures]] | ||
| + | == See Also == | ||
| + | |||
| + | * [http://en.wikipedia.org/wiki/Lysozyme Lysozyme] | ||
| + | * [http://en.wikipedia.org/wiki/Glycoside_hydrolase#Retaining_glycoside_hydrolases Retaining Glycoside Hydrolases] | ||
| + | *[[Molecular Playground/Lysozyme]] | ||
| + | *[[User:Judy Voet/Lysozyme]] | ||
| + | *[[Lysozyme (Arabic)]] | ||
| + | *[[Lysozyme (hebrew)]] | ||
| + | |||
| + | == References == | ||
<references/> | <references/> | ||
| + | |||
| + | [[Category:Topic Page]] | ||
| + | |||
| + | [[ar: Lysozyme (Arabic)]] | ||
| + | [[he: Lysozyme (hebrew)]] | ||
Current revision
Contents |
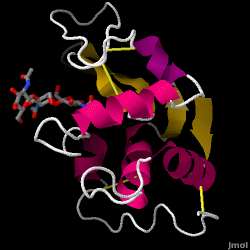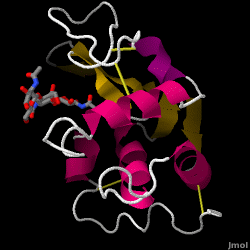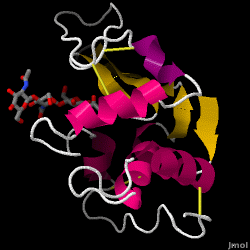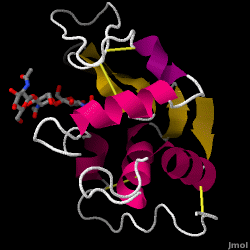
Lysozyme - also known as muramidase - is a powerful enzyme found in abundance in tears, saliva, and human milk. In humans, it is encoded in the LYZ gene. Since it is a small, easily available, and highly stable protein, it has been subject to extensive research regarding its function and structure.
Introduction
Lysozyme acts as a non-specific defense against bacteria and fungi. It is a component of the innate immune system, and is an important part of an infant's diet to ward off diarrhea. It is an enzyme known for its ability to degrade the polysaccharide architecture of many kinds of cell walls, normally for the purpose of protection against bacterial infection[1]. The structure of hen egg white (HEW) lysozyme, the focus of this article, is shown on the right. The antibacterial activity of hen egg white was first described by Laschtschenko in 1909[2]. It was characterized and named “lysozyme” by Alexander Fleming, the same person credited for the discovery of penicillin[3]. Discovery of the enzymatic activity was by accident; during the unrelated experiment, nasal drippings were inadvertently introduced to a petri dish containing a bacterial culture, which culture consequently exhibited the results of an as yet unknown enzymatic reaction. The observation of this unknown reaction led to further research on the components of this reaction as well as to the corresponding identification of the newfound "lysozyme." In 1965, David C. Phillips and coworkers determined the three-dimensional structure of lysozyme at 2 Å resolution [4]. Phillips' work was especially groundbreaking since Phillips had managed to successfully elucidate the structure of an enzyme via X-ray crystallography - a feat that had never before been accomplished[5][6]. Phillips' research also led to a structure-based hypothesis of its mechanism of action
[7].
Function
The primary function of lysozyme is to hydrolyze the bonds between the sugar molecules in the peptidoglycan layer of bacterial cell walls. Peptidoglycan is a rigid structure that provides support and protection to bacterial cells. By cleaving these bonds, lysozyme weakens the bacterial cell wall, leading to the rupture and death of the bacterium.
Lysozyme is particularly effective against Gram-positive bacteria, which have a relatively thick peptidoglycan layer. Gram-negative bacteria, on the other hand, have an outer membrane that provides additional protection, making them less susceptible to lysozyme.
In addition to its antimicrobial activity, lysozyme also plays a role in other physiological processes. It contributes to the maintenance of healthy epithelial tissues, such as those lining the respiratory and gastrointestinal tracts. Lysozyme helps prevent bacterial overgrowth and infection in these areas by inhibiting the growth of certain bacteria.
Lysozyme is widely distributed in nature and can be found in various organisms, including humans, animals, and plants. It is particularly abundant in the secretions of the lacrimal glands (tears) and salivary glands, where it helps protect the eyes and oral cavity from bacterial infections.
Due to its antimicrobial properties, lysozyme has been used in various applications, such as a natural food preservative and an additive in certain personal care products. It has also been studied for its potential therapeutic applications, including in the development of antimicrobial agents and the treatment of certain infections.
- Tail-associated lysozyme is expressed in bacteriophage T4 plays a role in digesting the peptidoglycan layering in lysis from within[8].
Chemical activity
The particular substrate of preference for this cleavage type is a (NAG-NAM)₃ hexasaccharide, within which substrate occurs the cleaving target glycosidic bond, NAM₄-β-O-NAG₅. The individual hexasaccharide binding units are designated A-F, with NAM₄-β-O-NAG₅ glycosidic bond cleavage preference corresponding to a D-E unit glycosidic bond cl
Lysozyme is known for damaging bacterial cell walls by catalyzing the hydrolysis of 1,4-beta-linkages between N-acetylmuramic acid (NAM) and N-acetyl-D-glucosamine (NAG) residues in peptidoglycan, and between N-acetyl-D-glucosamine residues in chitodextrins. In this way, lysozyme is efficient in lysing the cell walls of both bacteria and fungi. The location of cleavage for lysozyme on this architectural theme is the β(1-4) glycosidic linkage connecting the C1 carbon of NAM to the C4 carbon of NAG.
The particular substrate of preference for this cleavage type is a (NAG-NAM)₃ hexasaccharide, within which substrate occurs the cleaving target glycosidic bond, NAM₄-β-O-NAG₅. The individual hexasaccharide binding units are designated A-F, with NAM₄-β-O-NAG₅ glycosidic bond cleavage preference corresponding to a D-E unit glycosidic bond cleavage preference. Depending on the organism from which lysozyme is obtained, hydrolysis of the glycosidic bond proceeds with retention of configuration at the anomeric carbon (hen egg white) or with inversion (goose, phage T4).
Lysozyme efficiently acts on long (NAG-NAM) or (NAG) polymers. As the chain length gets smaller than six monomers, the catalytic rates drop substantially; in fact, trisaccharides act as competitive inhibitors by binding to the active site in a non-productive register.
| |||||||||||
Mechanism
Hydrolysis of glycosidic bonds by hen egg white lysozyme proceeds with retention of configuration. In 1953, Koshland [17] suggested that in general, retention of configuration implies a double-displacement mechanism (while inversion of configuration implies single displacement). For decades, two competing mechanistic hypotheses (Phillips[7]: dissociative mechanism with oxocarbenium intermediate; Koshland: two-step associative mechanism with covalent enzyme complex as intermediate) were considered, with data from 2001[15] tipping the scale toward the existence of a covalent intermediate. The absence of a substrate complex structure certainly contributed to difficulties distinguishing between possible mechanisms, as did the existence of two distinct mechanisms (retention and inversion of configuration) within the same structural family of enzymes (e.g. hen vs goose enzyme).
Lysozyme hydrolyzes a glycoside (hence the familial classification of lysozyme as a glycosylase[18]), which corresponds to the conversion of an acetal to a hemiacetal. The reaction proceeds in two steps as shown in the figure above. In the first step, Asp 52 acts as nucleophile and part of the sugar is the leaving group. In the second step, water acts as nucleophile and Asp 52 acts as leaving group. Both steps invert the configuration at the anomeric carbon, leading to an overall retention of configuration. Glu 35 acts as an acid in the first step (protonating the sugar the the glycosidic bond to make it a better electrophile) and as a base in the second step (deprotonating water to make it a better nucleophile). While the figure shows some of the sugars in a boat conformation to emphasize the inversion of configuration, these are not observed experimentally but are rather found in a chair conformation.
Applications of Lysozyme
Since lysozyme has been widely recognized for its antibacterial and antifungal properties, it has a wide variety of uses both in biochemical and pharmaceutical applications. In molecular biology, lysozyme is often used in the alkaline-lysis procedure for extracting and isolating plasmid DNA. It is used extensively in the pharmaceutical field for destroying gram-positive bacteria, and can be used to support already-existing immune defenses to fight bacterial infections. This enzyme is particularly important for preventing bacterial diseases in infants. Because of its antibacterial properties, lysozyme can also be used in the food industry to help prevent spoilage of foods.
3D structures of lysozome
See Also
- Lysozyme
- Retaining Glycoside Hydrolases
- Molecular Playground/Lysozyme
- User:Judy Voet/Lysozyme
- Lysozyme (Arabic)
- Lysozyme (hebrew)
References
- ↑ Ragland SA, Criss AK. From bacterial killing to immune modulation: Recent insights into the functions of lysozyme. PLoS Pathog. 2017 Sep 21;13(9):e1006512. doi: 10.1371/journal.ppat.1006512., eCollection 2017 Sep. PMID:28934357 doi:http://dx.doi.org/10.1371/journal.ppat.1006512
- ↑ Laschtschenko,P. (1909) Über die keimtötende und entwicklungshemmende Wirkung von Hühnereiweiss. Z. Hyg. Infektionskrankh.,64,419-427.
- ↑ Fleming, A. (1922) On a remarkable bacteriolytic element found in tissues and secretions. Proc.Roy.Soc.(London),93,306-317.
- ↑ Blake CC, Koenig DF, Mair GA, North AC, Phillips DC, Sarma VR. Structure of hen egg-white lysozyme. A three-dimensional Fourier synthesis at 2 Angstrom resolution. Nature. 1965 May 22;206(986):757-61. PMID:5891407
- ↑ Bugg, T. 1997. An Introduction to Enzyme and Coenzyme Chemistry. Blackwell Science Ltd., Oxford
- ↑ Earliest Solutions for Macromolecular Crystal Structures.
- ↑ 7.0 7.1 Blake CC, Johnson LN, Mair GA, North AC, Phillips DC, Sarma VR. Crystallographic studies of the activity of hen egg-white lysozyme. Proc R Soc Lond B Biol Sci. 1967 Apr 18;167(1009):378-88. doi:, 10.1098/rspb.1967.0035. PMID:4382801 doi:http://dx.doi.org/10.1098/rspb.1967.0035
- ↑ Nakagawa H, Arisaka F, Ishii S. Isolation and characterization of the bacteriophage T4 tail-associated lysozyme. J Virol. 1985 May;54(2):460-6. PMID:3157805 doi:10.1128/JVI.54.2.460-466.1985
- ↑ Image from: http://www.vuw.ac.nz/staff/paul_teesdale-spittle/essentials/chapter-6/proteins/lysozyme.htm
- ↑ Richardson JS. Early ribbon drawings of proteins. Nat Struct Biol. 2000 Aug;7(8):624-5. doi: 10.1038/77912. PMID:10932243 doi:http://dx.doi.org/10.1038/77912
- ↑ http://mcdb-webarchive.mcdb.ucsb.edu/sears/biochemistry/tw-enz/lysozyme/HEWL/lysozyme-overview.htm
- ↑ Blake CC, Koenig DF, Mair GA, North AC, Phillips DC, Sarma VR. Structure of hen egg-white lysozyme. A three-dimensional Fourier synthesis at 2 Angstrom resolution. Nature. 1965 May 22;206(986):757-61. PMID:5891407
- ↑ Johnson LN, Phillips DC. Structure of some crystalline lysozyme-inhibitor complexes determined by X-ray analysis at 6 Angstrom resolution. Nature. 1965 May 22;206(986):761-3. PMID:5840126
- ↑ Phillips (1966) Scientific American 215, 76-90
- ↑ 15.0 15.1 Vocadlo DJ, Davies GJ, Laine R, Withers SG. Catalysis by hen egg-white lysozyme proceeds via a covalent intermediate. Nature. 2001 Aug 23;412(6849):835-8. PMID:11518970 doi:10.1038/35090602
- ↑ Davies, Withers and Vocadlo (2009) The Chitopentaose Complex of a Mutant Hen Egg-White Lysozyme Displays No Distortion of the –1 Sugar Away from a 4C1 Chair Conformation, Australian Journal of Chemistry 62(6) 528-532
- ↑ Koshland, D. E. (1953). Biol. Rev. 28, 416–436
- ↑ https://bio.libretexts.org/Bookshelves/Biochemistry/Book%3A_Biochemistry_Online_(Jakubowski)/07%3A_CATALYSIS/B._Mechanisms_of_Enzyme-Catalyzed_Reactions/B2.__Lysozyme
Proteopedia Page Contributors and Editors (what is this?)
Karsten Theis, Michal Harel, Anne Goodling, Alexander Berchansky, Eric Martz, Joel L. Sussman, Karl Oberholser, Jaime Prilusky, John Ripollone, Daniel Kreider, Judy Voet, John S. de Banzie