AppA protein BLUF domain
From Proteopedia
(Difference between revisions)
(New page: {{STRUCTURE_1x0p| PDB=1x0p | SCENE= }} =Introduction= The proteins containing sensors for blue light using [http://en.wikipedia.org/wiki/Flavin_adenine_dinucleotide FAD] (BLUF) domains are...) |
m (Appa protein BLUF domain moved to AppA protein BLUF domain) |
||
| (21 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='1x0p' size='350' side='right' caption='Structure of BLUF domain protein complex with FAD (PDB code [[1x0p]]).' scene=''> | |
=Introduction= | =Introduction= | ||
| - | + | '''AppA''' is an anti repressor in which blue light excitation induces a long-lived red-shift absorbance in photosystem synthesis<ref>PMID:12230978</ref> . AppA contains sensor for '''blue light using''' [http://en.wikipedia.org/wiki/Flavin_adenine_dinucleotide FAD] '''(BLUF) domains''' are one class of photoreceptor family that utilizes a flavin [http://en.wikipedia.org/wiki/Chromophore chromophore]<ref name ="one">PMID: 15876364</ref>. The other two classes include [http://en.wikipedia.org/wiki/Phototropin phototropins] (LOV) and [http://en.wikipedia.org/wiki/Cryptochrome cryptochromes]<ref name="two">PMID: 14730990</ref>. The BLUF domain was first discovered in [http://microbewiki.kenyon.edu/index.php/Rhodobacter ''Rhodobacter sphaeroides''] as the blue light photoreceptor involved in the repression of photosynthesis genes in '''AppA''' protein<ref name="one" /><ref name ="three">PMID: 12230978</ref>. The BLUF domain is known to exist in many bacteria, including cyanobacteria. | |
One unique photosensing property of BLUF domain is a light induced spectral shift in the flavin absorption spectrum where the wavelength is longer by approximately 10nm<ref name="one" />. In AppA this shift occurs quickly upon illumination and is slowly reversed upon return to darkness. This spectral shift is not well understood and is not observed in other flavin binding photoreceptors. There have been two proposed models to explain the spectral shift observed in BLUF containing proteins: π-π stacking between the isoalloxazine ring of flavin and the phenol sidechain of a conserved tyrosine residue or protonation and deprotonation of the flavin ring coupled directly or indirectly with the conserved tyrosine residue<ref name ="four">PMID: 14556317</ref><ref name ="five">PMID: 15659451</ref>. These two models are based upon the conserved tyrosine residue Tyr9 in the T110078 protein. | One unique photosensing property of BLUF domain is a light induced spectral shift in the flavin absorption spectrum where the wavelength is longer by approximately 10nm<ref name="one" />. In AppA this shift occurs quickly upon illumination and is slowly reversed upon return to darkness. This spectral shift is not well understood and is not observed in other flavin binding photoreceptors. There have been two proposed models to explain the spectral shift observed in BLUF containing proteins: π-π stacking between the isoalloxazine ring of flavin and the phenol sidechain of a conserved tyrosine residue or protonation and deprotonation of the flavin ring coupled directly or indirectly with the conserved tyrosine residue<ref name ="four">PMID: 14556317</ref><ref name ="five">PMID: 15659451</ref>. These two models are based upon the conserved tyrosine residue Tyr9 in the T110078 protein. | ||
| Line 10: | Line 10: | ||
==Domains== | ==Domains== | ||
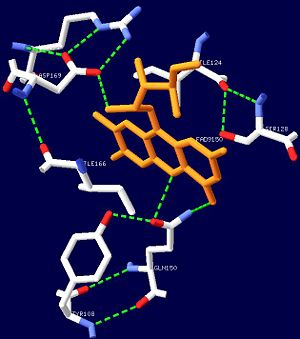
[[Image:FADbonding.jpg|thumb|left|300px|Figure 1. The hydrogen bonds the FAD ligand forms with nearby amino acid residues.]] | [[Image:FADbonding.jpg|thumb|left|300px|Figure 1. The hydrogen bonds the FAD ligand forms with nearby amino acid residues.]] | ||
| - | The BLUF domain is a <scene name=' | + | The BLUF domain is a <scene name='44/447813/Cv/15'>decamer</scene> with a molecular weight of approximately 160kDa<ref name="one" />. There are ten monomers observed in each asymmetric unit. The crystalline structure of the BLUF domain from the T110078 protein was solved by single isomorphous replacement (SIR) method using a mercury derivative. The <scene name='44/447813/Cv/17'>double ringed decamer has a diameter of approximately 95Å</scene>, a <scene name='44/447813/Cv/18'>thickness of 60Å</scene> and a <scene name='44/447813/Cv/19'>central channel approximately 35Å in diameter</scene><ref name="one" />. |
| - | Each monomer is comprised of 5 <scene name=' | + | Each monomer is comprised of 5 <scene name='44/447813/Cv/12'>β-strands</scene> and 4 <scene name='44/447813/Cv/13'>α-helices</scene> in the order of β1α1β2β3α2β4β5α3α4. Specifically, the BLUF domain of the monomer contains β1α1β2β3α2β4β5, while the C-terminal domain contains α3α4<ref name="one" />. The C-terminal domain interacts with the end of the β-sheet of the neighbouring monomer. |
| - | The isoalloxazine ring of FAD is located between <scene name=' | + | The isoalloxazine ring of FAD is located between <scene name='44/447813/Cv/14'>α1 and α2</scene> of the BLUF domain, between <scene name='44/447813/Cv/4'>two highly conserved hydrophobic residues: Ile124 and Ile166</scene><ref name="one" />. FAD forms hydrogen bonds with the following amino acid residues: <scene name='44/447813/Cv/5'>Asn131, Asn132, Gln150, Arg165 and Asp169</scene> (see also static image Figure 1)<ref name="one" />. More specifically, the <scene name='44/447813/Cv/6'>side chain of Asn131 binds to O2 of FAD</scene> and <scene name='44/447813/Cv/7'>Asn132 binds to N3 and O4 of FAD</scene>. The <scene name='44/447813/Cv/8'>guanido group of Arg165 contributes to a network between FAD and the apo protein</scene>. The amide N of the Gln150 sidechain interacts with N5 and O4 of FAD through hydrogen bonding, while the amide O of the sidechain is closely linked with the hydroxyl oxygen of the highly conserved Tyr8 residue, forming a <scene name='44/447813/Cv/9'>FAD-Gln150-Tyr108 network</scene><ref name="one" />. This conserved Tyr108 residue is the only residue that has been shown to be essential for light reaction in the BLUF domain containing AppA and Slr1694 proteins<ref name ="seven">PMID: 17042486</ref>. |
| + | *<scene name='44/447813/Cv/11'>Whole FAD binding site</scene>. | ||
==Further Analyses== | ==Further Analyses== | ||
| - | While | + | While Asn132, Gln150, As169, Arg171, His172 and Ser110 are completely conserved residues, Asn131, Arg165 and Ser128 are only moderately conserved. This network surrounding FAD is thought to be highly conserved in all BLUF domains. In order to study the importance of the interactions between the isoalloxaizine ring of FAD, and Asn131, Asn132 and Gln150, the amino acids were replaced with alanine residues<ref name="one" />. It was observed that the spectra of the mutants were qualitatively similar to that of the wild-type. However, the low energy absorbance peak of N132A (Asn132 replaced with Ala) was blue shifted compared to the wildtype by 6nm<ref name="one" />. This blue shift can be accounted for by the lack of hydrogen bonding between the side-chain and isoalloxazine ring<ref name="one" />. In the wild-type protein, Asn132 forms 2 hydrogen bonds with FAD; the absence of these bonds allows relocation of an electron on FAD, resulting on the spectral blue shift. |
| - | The | + | The Q140A mutant showed very little spectral changes during illumination. These results suggest that Gln150 is critical for light reaction in BLUF domains and is thus totally conserved<ref name="one" />. Transient bleaching of flavin was observed instead of red-shifting; this suggests that light excitation of flavin in Q150A resulted in photoreduction of flavin<ref name="one" />. In general, a free flavin in solution is sequentially reduced with two electrons upon exposure to light. The first electron reduces the double bond at N5 and the second electron reduces at N1 of the isoalloxazine ring. In BLUF proteins, the role of Gln50 may be to prevent the initial photoreduction at N5 and allow a reaction leading to the spectral shift to a longer wavelength. This may be achieved by the hydrogen bonding between the amide N of Gln50 and the N5 of the isoalloxazine ring of FAD. |
Similar hydrogen bonding networks surrounding the isoalloxazine ring are found in other light sensing proteins such as the LOV proteins<ref name="one" />. The most critical residue in the flavin sensor proteins appears to be the one that interacts closely with N5 of the isoalloxazine ring<ref name="one" />. | Similar hydrogen bonding networks surrounding the isoalloxazine ring are found in other light sensing proteins such as the LOV proteins<ref name="one" />. The most critical residue in the flavin sensor proteins appears to be the one that interacts closely with N5 of the isoalloxazine ring<ref name="one" />. | ||
| Line 24: | Line 25: | ||
=Function= | =Function= | ||
Overall, the main function of the BLUF domain is to detect and respond to blue light. More specifically, in ''R. sphaeroides'', the BLUF domain is a blue light photo receptor involved in repressing the photosynthesis genes at the N-terminal region of the AppA protein<ref name="three" />. In ''E. gracilis'', the BLUF domain of PAC complexes serves as a blue light receptor in photophobic responses<ref name="six" />. | Overall, the main function of the BLUF domain is to detect and respond to blue light. More specifically, in ''R. sphaeroides'', the BLUF domain is a blue light photo receptor involved in repressing the photosynthesis genes at the N-terminal region of the AppA protein<ref name="three" />. In ''E. gracilis'', the BLUF domain of PAC complexes serves as a blue light receptor in photophobic responses<ref name="six" />. | ||
| + | </StructureSection> | ||
| + | =3D structures of AppA= | ||
| + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | |||
| + | [[2byc]] – RsBlue light receptor BLUF + FMN - ''Rhodobacter sphaeroides''<br /> | ||
| + | [[2bun]] – RsAppA BLUF domain + FAD – NMR <br /> | ||
| + | [[2iyg]], [[2iyi]] – RsAppA BLUF domain (mutant) + FMN <br /> | ||
| + | [[4heh]] – RsAppA SCHIC domain <br /> | ||
| + | [[4hh3]] – RsAppA SCHIC domain (mutant) + PpsR<br /> | ||
| + | [[4hh1]] – RsAppA + FMN<br /> | ||
| + | [[4hh0]] – RsAppA (mutant) + FMN<br /> | ||
| + | [[1x0p]] - BLUF + FAD - ''Thermosynechococcus elongatus''<br /> | ||
| + | [[1xoc]] – AppA + peptide - ''Bacillus subtilis''<br /> | ||
| + | [[4tsr]] – AppA (mutant) + inositol hexasulfate - ''Eschericia coli''<br /> | ||
=References= | =References= | ||
<references /> | <references /> | ||
| + | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
3D structures of AppA
Updated on 13-May-2021
2byc – RsBlue light receptor BLUF + FMN - Rhodobacter sphaeroides
2bun – RsAppA BLUF domain + FAD – NMR
2iyg, 2iyi – RsAppA BLUF domain (mutant) + FMN
4heh – RsAppA SCHIC domain
4hh3 – RsAppA SCHIC domain (mutant) + PpsR
4hh1 – RsAppA + FMN
4hh0 – RsAppA (mutant) + FMN
1x0p - BLUF + FAD - Thermosynechococcus elongatus
1xoc – AppA + peptide - Bacillus subtilis
4tsr – AppA (mutant) + inositol hexasulfate - Eschericia coli
References
- ↑ Masuda S, Bauer CE. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell. 2002 Sep 6;110(5):613-23. PMID:12230978
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 2.16 2.17 Kita A, Okajima K, Morimoto Y, Ikeuchi M, Miki K. Structure of a cyanobacterial BLUF protein, Tll0078, containing a novel FAD-binding blue light sensor domain. J Mol Biol. 2005 May 27;349(1):1-9. Epub 2005 Apr 9. PMID:15876364 doi:10.1016/j.jmb.2005.03.067
- ↑ van der Horst MA, Hellingwerf KJ. Photoreceptor proteins, "star actors of modern times": a review of the functional dynamics in the structure of representative members of six different photoreceptor families. Acc Chem Res. 2004 Jan;37(1):13-20. PMID:14730990 doi:10.1021/ar020219d
- ↑ 4.0 4.1 Masuda S, Bauer CE. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell. 2002 Sep 6;110(5):613-23. PMID:12230978
- ↑ Laan W, van der Horst MA, van Stokkum IH, Hellingwerf KJ. Initial characterization of the primary photochemistry of AppA, a blue-light-using flavin adenine dinucleotide-domain containing transcriptional antirepressor protein from Rhodobacter sphaeroides: a key role for reversible intramolecular proton transfer from the flavin adenine dinucleotide chromophore to a conserved tyrosine? Photochem Photobiol. 2003 Sep;78(3):290-7. PMID:14556317
- ↑ Hasegawa K, Masuda S, Ono TA. Spectroscopic analysis of the dark relaxation process of a photocycle in a sensor of blue light using FAD (BLUF) protein Slr1694 of the cyanobacterium Synechocystis sp. PCC6803. Plant Cell Physiol. 2005 Jan;46(1):136-46. Epub 2005 Jan 19. PMID:15659451 doi:10.1093/pcp/pci003
- ↑ 7.0 7.1 Iseki M, Matsunaga S, Murakami A, Ohno K, Shiga K, Yoshida K, Sugai M, Takahashi T, Hori T, Watanabe M. A blue-light-activated adenylyl cyclase mediates photoavoidance in Euglena gracilis. Nature. 2002 Feb 28;415(6875):1047-51. PMID:11875575 doi:10.1038/4151047a
- ↑ Yuan H, Anderson S, Masuda S, Dragnea V, Moffat K, Bauer C. Crystal structures of the Synechocystis photoreceptor Slr1694 reveal distinct structural states related to signaling. Biochemistry. 2006 Oct 24;45(42):12687-94. PMID:17042486 doi:10.1021/bi061435n
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Amanda Cookhouse, Jaime Prilusky