Ketosteroid Isomerase
From Proteopedia
(Difference between revisions)
(→Protein Folding) |
|||
| (30 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
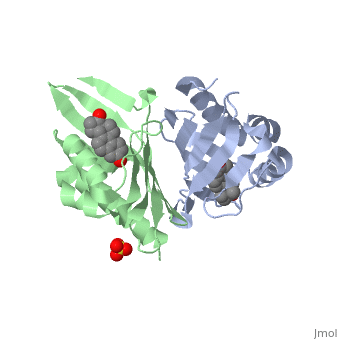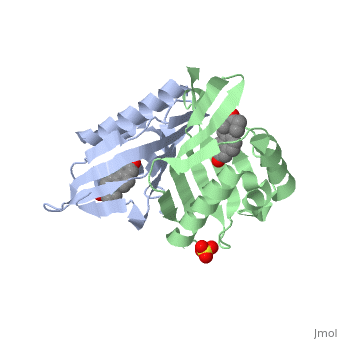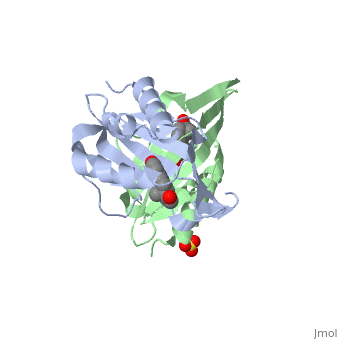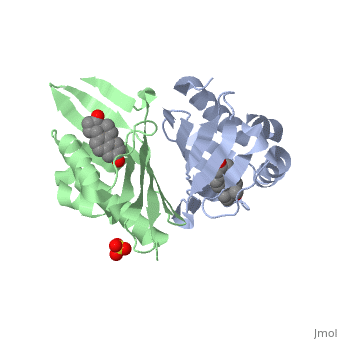
| - | == | + | <StructureSection load='1qjg' size='350' side='right' scene='' caption='Ketosteroid isomerase complex with transition state analog equilenin and sulfate (PDB code [[1qjg]])'> |
| - | ===Introduction | + | ==Introduction== |
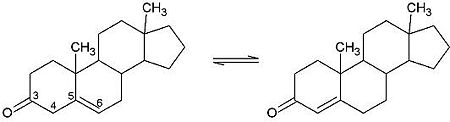
| - | + | <scene name='User:Laura_M._Haynes/Sandbox_1/Ksi/4'>Ketosteroid isomerase</scene> (KSI, EC#5.3.3.1) or '''steroid delta-isomerase''' is an enzyme that catalyzes the isomerization of 3-oxo-Δ<sup>5</sup> ketosteroids to their hormonally active Δ<sup>4</sup>-conjugated isomers, as illustrated below.<ref name="Pollack">PMID:15381400</ref>, <ref name="Talalay">PMID:13276386 </ref> | |
| - | <scene name='User:Laura_M._Haynes/Sandbox_1/Ksi/4'>Ketosteroid isomerase</scene> (KSI, EC#5.3.3.1) is an enzyme that catalyzes the isomerization of 3-oxo-Δ<sup>5</sup> ketosteroids to their hormonally active Δ<sup>4</sup>-conjugated isomers, as illustrated below.<ref name="Pollack">PMID:15381400</ref>, <ref name="Talalay">PMID:13276386 </ref> | + | |
| - | [[Image:Reaction.jpg| | + | [[Image:Reaction.jpg|left|450px|thumb]] |
| - | This reaction is essential in the biosynthesis of steroids in mammals where KSI is a membrane-bound complex.<ref name="Ha">PMID:11751047</ref> In bacteria, however, KSI exists as a soluble protein is involves in catabolism of steroids.<ref name="Ha" /> It was first isolated in and has been extensively studied in [http://en.wikipedia.org/wiki/Comamonas_testosteroni ''Commamonas tetosteroni''] (TI), a bacteria that is capable of growth with testosterone as its sole carbon source.<ref name="Stanier1966">PMID:11751047</ref> Structural and kinetic studies of this and its homolog from [http://en.wikipedia.org/wiki/Pseudomonas_putida ''Pseudomonas putida''] with which it shares 34% sequence and near identical structural homology.<ref name="Pollack" />,<ref name="Ha" /> It is one of the most efficient known enzymes with an essentially diffusion limited rate of catalysis.<ref name="Talalay" />,<ref name="Wu">PMID:9103200 </ref> It is capable of increasing the catalytic rate by eleven orders of magnitude.<ref name="Murzin">PMID:9666335 </ref> The high degree of efficiency is believed to be due to a preference for the transition state to move towards products rather than reactants although the exact mechanism of this preference is unclear.<ref name="Pollack" /> Its high catalytic efficiency and unique active site geometry have made it fertile ground for examining the validity of the [http://en.wikipedia.org/wiki/Low-barrier_hydrogen_bond low barrier hydrogen bond] hypothesis<ref name="Cleland1998">PMID:9748211 </ref> and electrostatic preorganization.<ref name="Kraut2006">PMID:16602823 </ref>. | + | This reaction is essential in the biosynthesis of steroids in mammals where KSI is a membrane-bound complex.<ref name="Ha">PMID:11751047</ref> In bacteria, however, KSI exists as a soluble protein is involves in catabolism of steroids.<ref name="Ha" /> It was first isolated in and has been extensively studied in [http://en.wikipedia.org/wiki/Comamonas_testosteroni ''Commamonas tetosteroni''] (TI), a bacteria that is capable of growth with testosterone as its sole carbon source.<ref name="Stanier1966">PMID:11751047</ref> Structural and kinetic studies of this and its homolog from [http://en.wikipedia.org/wiki/Pseudomonas_putida ''Pseudomonas putida''] with which it shares 34% sequence and near identical structural homology.<ref name="Pollack" />,<ref name="Ha" /> It is one of the most efficient known enzymes with an essentially diffusion limited rate of catalysis.<ref name="Talalay" />,<ref name="Wu">PMID:9103200 </ref> It is capable of increasing the catalytic rate by eleven orders of magnitude.<ref name="Murzin">PMID:9666335 </ref> The high degree of efficiency is believed to be due to a preference for the transition state to move towards products rather than reactants although the exact mechanism of this preference is unclear.<ref name="Pollack" /> Its high catalytic efficiency and unique active site geometry have made it fertile ground for examining the validity of the [http://en.wikipedia.org/wiki/Low-barrier_hydrogen_bond low barrier hydrogen bond] hypothesis<ref name="Cleland1998">PMID:9748211 </ref> and electrostatic preorganization.<ref name="Kraut2006">PMID:16602823 </ref>. See also [[Isomerases]]. |
==Structure== | ==Structure== | ||
| Line 13: | Line 12: | ||
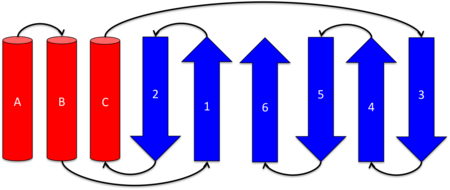
| - | [[Image:top2.png|thumb| | + | [[Image:top2.png|thumb|left|450px|'''Topology diagram of KSI's structure''' Helices A, B, and C are in an antiparallel arrangement. The six β-strands form a mixed β-sheet.]] |
===Alpha-Helices=== | ===Alpha-Helices=== | ||
| Line 22: | Line 21: | ||
α-helix capping motifs are defined by specific patterns of hydrophobic interactions and hydrogen bonding that occur at both the initiation and termination of these secondary structural elements. <ref name="Aurora">PMID:9514257 </ref> Capping motifs can also help support the formation of tertiary structural elements. KSI contains examples several of these motifs involved in both C-terminal and N-teminal capping, with two illustrations of C-terminal capping motifs as illustrated below (nomenclature adapted from that of Aurora and Rose <ref name="Aurora" />): | α-helix capping motifs are defined by specific patterns of hydrophobic interactions and hydrogen bonding that occur at both the initiation and termination of these secondary structural elements. <ref name="Aurora">PMID:9514257 </ref> Capping motifs can also help support the formation of tertiary structural elements. KSI contains examples several of these motifs involved in both C-terminal and N-teminal capping, with two illustrations of C-terminal capping motifs as illustrated below (nomenclature adapted from that of Aurora and Rose <ref name="Aurora" />): | ||
| - | [[Image: helix_capping_motifs.png|thumb| | + | [[Image: helix_capping_motifs.png|thumb|left|450px|'''Examples of C-terminal helix capping motifs present in KSI.''' (A) α-L capping motif. (B) Proline capping motif.]] |
=====Alpha-L C-terminal capping motif===== | =====Alpha-L C-terminal capping motif===== | ||
| Line 43: | Line 42: | ||
The biologically active unit of KSI is a 2-fold symmetric dimer in which the two chains are packed together via hydrophobic and electrostatic interactions between the "back faces" of the β-sheets. The curved β-sheets on each of the monomers expose convex faces to each other, forming well-defined interactions with each monomer having a self-contained active site.<ref name="folding">PMID:11305917</ref> In their NMR structure of KSI, Massiah et al. <ref name="Massiah">PMID:9778345</ref> identified interchain hydrophobic interactions (A), as well, a number of polar residues (B) located within the dimer interface. | The biologically active unit of KSI is a 2-fold symmetric dimer in which the two chains are packed together via hydrophobic and electrostatic interactions between the "back faces" of the β-sheets. The curved β-sheets on each of the monomers expose convex faces to each other, forming well-defined interactions with each monomer having a self-contained active site.<ref name="folding">PMID:11305917</ref> In their NMR structure of KSI, Massiah et al. <ref name="Massiah">PMID:9778345</ref> identified interchain hydrophobic interactions (A), as well, a number of polar residues (B) located within the dimer interface. | ||
| - | [[Image:dimer_interface.png|thumb| | + | [[Image:dimer_interface.png|thumb|left|450px|'''Interactions at KSI's dimer interface''' (A) Hydrophobic residues. (B) Hydrophillic residues.]] |
[[Image:Kim1.png|thumb|right|175px|'''Water mediated dimer interaction between Thr68 and Arg72.''' ]] In their X-ray crystal structure of KSI, Kim et al.<ref name="1OHO">PMID:9369474 </ref> identified bound water molecules within the dimer interface which may mediate hydrogen-bonds between Thr68, Arg72, and Asp96. The water mediated hydrogen bond between Thr68 and Arg72 is illustrated below. Kim et al. also identified possibe interchain hydrogen bonds between the backbone carbonyl oxygens of Val71, Ala73, and Val97 with the side chains of Asn120, Ser117, and Arg72 respectively based on the solution structure of Wu et al.<ref name="Wu" />. An intersidechain hydrogen-bond is also suspected between His75 and Gln118. | [[Image:Kim1.png|thumb|right|175px|'''Water mediated dimer interaction between Thr68 and Arg72.''' ]] In their X-ray crystal structure of KSI, Kim et al.<ref name="1OHO">PMID:9369474 </ref> identified bound water molecules within the dimer interface which may mediate hydrogen-bonds between Thr68, Arg72, and Asp96. The water mediated hydrogen bond between Thr68 and Arg72 is illustrated below. Kim et al. also identified possibe interchain hydrogen bonds between the backbone carbonyl oxygens of Val71, Ala73, and Val97 with the side chains of Asn120, Ser117, and Arg72 respectively based on the solution structure of Wu et al.<ref name="Wu" />. An intersidechain hydrogen-bond is also suspected between His75 and Gln118. | ||
| Line 51: | Line 50: | ||
KSI's active site is located within a hydrophobic cavity formed helices B and C crossing over the "front face" of the β-sheet that is approximately 8.5 by 9.5 Å at its opening and is 16 Å deep.<ref name="Wu" /> The cavity is lined with <scene name='User:Laura_M._Haynes/Sandbox_1/Beta-phobics/1'>hydrophobic "front facing" residues</scene> from the β-sheet(Val36, Pro39, Leu63, Val65, Leu67, Val71, Phe80, Phe82, Val84, Val95, Pro97, Phe101, Ala114, and Phe116). <scene name='User:Laura_M._Haynes/Sandbox_1/Helix-phobic/1'>Hydrophobic residues on the other side of the cavity</scene> contributed from the α-helices include: Val11, Tyr14, Val15, Leu18, Phe54, and Tyr55.<ref name="Wu" /> | KSI's active site is located within a hydrophobic cavity formed helices B and C crossing over the "front face" of the β-sheet that is approximately 8.5 by 9.5 Å at its opening and is 16 Å deep.<ref name="Wu" /> The cavity is lined with <scene name='User:Laura_M._Haynes/Sandbox_1/Beta-phobics/1'>hydrophobic "front facing" residues</scene> from the β-sheet(Val36, Pro39, Leu63, Val65, Leu67, Val71, Phe80, Phe82, Val84, Val95, Pro97, Phe101, Ala114, and Phe116). <scene name='User:Laura_M._Haynes/Sandbox_1/Helix-phobic/1'>Hydrophobic residues on the other side of the cavity</scene> contributed from the α-helices include: Val11, Tyr14, Val15, Leu18, Phe54, and Tyr55.<ref name="Wu" /> | ||
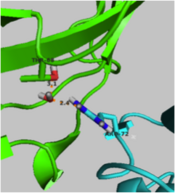
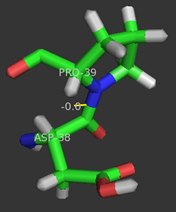
| - | Although the <scene name='User:Laura_M._Haynes/Sandbox_1/Active_site_1isk/1'>active site</scene> of KSI is notably hydrophobic, it contains several hydrophilic residues believed to be important to the enzymatic function of the protein. The hydrophobic active site of KSI contains an aspartate residue at position 99 and a tyrosine residue at position 14 (according to the numbering for the ''Commamonas tetosteroni'' protein, which will be used throughout) that are capable of forming hydrogen bonds with the 3-position carbonyl of the steroid and form an active site oxyanion hole.<ref name="Pollack" />,<ref name="Sigala2008">PMID:18808119 </ref> Additionally, the active site contains an aspartate residue at position 38 that is participates in the catalytic activity of KSI.<ref name="Pollack" /> [[Image:cis.png|thumb| | + | Although the <scene name='User:Laura_M._Haynes/Sandbox_1/Active_site_1isk/1'>active site</scene> of KSI is notably hydrophobic, it contains several hydrophilic residues believed to be important to the enzymatic function of the protein. The hydrophobic active site of KSI contains an aspartate residue at position 99 and a tyrosine residue at position 14 (according to the numbering for the ''Commamonas tetosteroni'' protein, which will be used throughout) that are capable of forming hydrogen bonds with the 3-position carbonyl of the steroid and form an active site oxyanion hole.<ref name="Pollack" />,<ref name="Sigala2008">PMID:18808119 </ref> Additionally, the active site contains an aspartate residue at position 38 that is participates in the catalytic activity of KSI.<ref name="Pollack" /> [[Image:cis.png|thumb|left|'''Cis-Pro39.''']] |
| - | Upon substrate binding the the three α-helices become more tightly packed with the "front face" of the β-sheet. This in turn allows Tyr14 to approach Asp99 and the substrate. | + | <html5media height=“315” width=“560”>https://www.youtube.com/embed/o6XSjDNyGdw</html5media> |
| - | + | ||
| + | <br>Upon substrate binding the the three α-helices become more tightly packed with the "front face" of the β-sheet. This in turn allows Tyr14 to approach Asp99 and the substrate. | ||
===Cis-Peptide Bond=== | ===Cis-Peptide Bond=== | ||
| Line 62: | Line 62: | ||
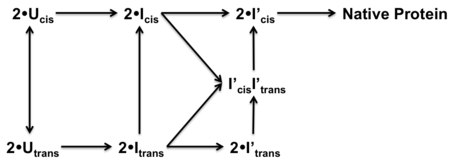
The work of Kim et al. suggests that KSI assembles into its biologically active dimer structure via a multistep pathway.<ref name="folding" /> The monomers initially exist in an unfolded configuration (U) with Pro39 in a trans configuration. The monomers then fold into an intermediate configuration that is capable of binding transition state analogs (I). At this stage the native secondary structures have most likely formed, however, it is also likely that hydrophobic residues have been internalized into the core. The tertiary structure formation is most likely promoted by the dimerized intermediate structure (I') before folding to the final native structure. Cis-trans isomerization is the rate-limiting step with dimer structures being formed preferentially from cis-Pro39 monomers. | The work of Kim et al. suggests that KSI assembles into its biologically active dimer structure via a multistep pathway.<ref name="folding" /> The monomers initially exist in an unfolded configuration (U) with Pro39 in a trans configuration. The monomers then fold into an intermediate configuration that is capable of binding transition state analogs (I). At this stage the native secondary structures have most likely formed, however, it is also likely that hydrophobic residues have been internalized into the core. The tertiary structure formation is most likely promoted by the dimerized intermediate structure (I') before folding to the final native structure. Cis-trans isomerization is the rate-limiting step with dimer structures being formed preferentially from cis-Pro39 monomers. | ||
| - | [[Image:folding.png|thumb|center| | + | [[Image:folding.png|thumb|center|450px|'''Protein folding pathway of KSI.''' (U) Unfolded protein. (I) Monomer intermediate. (I') Dimerized intermediate.]] |
==Enzymology== | ==Enzymology== | ||
| - | < | + | <scene name='User:Laura_M._Haynes/Sandbox_1/1qjg/1'>A monomer of ketosteroid isomerase is shown in complex with the transition state analog</scene> ([http://en.wikipedia.org/wiki/Equilenin equinelin]; PBD Structure ID [[1qjg|1QJG]]) |
| + | |||
===General Mechanism=== | ===General Mechanism=== | ||
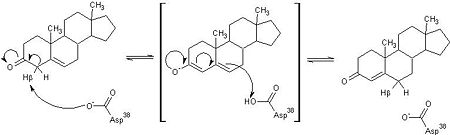
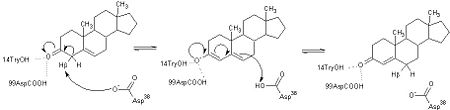
The general mechanism of the proposed reaction of ketosteroid isomerase involves the breaking of a C-H bond adjacent to a carbonyl. This is typically regarded as a difficult reaction due to instability of the intermediate; however it is observed in a number of enzyme-mediated biological reactions.<ref name="Pollack" /> In line with other biological reactions, the mechanism of KSI involves the abstraction the β-hyrdogen from the 4-position carbon resulting in the formation of an enol intermediate, which is followed by reketonization.<ref name="Pollack" />,<ref name="Ha" /> Structural and kinetic studies suggest that Asp<sup>38</sup> (numbering is that of the TI varient of KSI) serves as a general base in this reaction as shown below and abstracts the β-proton from C4 with the ''syn'' orbitals of its carboxylate group.<ref name="Wu" /> Note the formation of the unstable enolate intermediate. | The general mechanism of the proposed reaction of ketosteroid isomerase involves the breaking of a C-H bond adjacent to a carbonyl. This is typically regarded as a difficult reaction due to instability of the intermediate; however it is observed in a number of enzyme-mediated biological reactions.<ref name="Pollack" /> In line with other biological reactions, the mechanism of KSI involves the abstraction the β-hyrdogen from the 4-position carbon resulting in the formation of an enol intermediate, which is followed by reketonization.<ref name="Pollack" />,<ref name="Ha" /> Structural and kinetic studies suggest that Asp<sup>38</sup> (numbering is that of the TI varient of KSI) serves as a general base in this reaction as shown below and abstracts the β-proton from C4 with the ''syn'' orbitals of its carboxylate group.<ref name="Wu" /> Note the formation of the unstable enolate intermediate. | ||
| - | [[Image:General_base.jpg| | + | [[Image:General_base.jpg|left|450px|thumb]] |
Tyr<sup>14</sup> and Asp<sup>99</sup> are believed to participate in hydrogen bonding to the O-3 carbonyl of the substrate steroid and stabilize reaction intermediates. Tyr<sup>14</sup> is also believed to participate in a low barrier hydrogen bond with the 3-position oxygen of the steroid, thereby facilitating the abstraction of the β-hydrogen at the 4-position. There are two proposed models of this hydrogen bonding. In <scene name='User:Laura_M._Haynes/Sandbox_1/Model_1_1qjg/1'>Model 1</scene>, Tyr<sup>14</sup> and Asp<sup>99</sup> are both bound to the 3-position oxygen, whereas, in <scene name='User:Laura_M._Haynes/Sandbox_1/Model_2_1qjg/1'>Model 2</scene>, they form a hydrogen bonding network. These are <scene name='User:Laura_M._Haynes/Sandbox_1/1qjg/1'>shown here</scene> as a single monomer in complex with the intermediate analog [http://en.wikipedia.org/wiki/Equilenin equilenin].<ref name="Cho2">PMID:10551849</ref> | Tyr<sup>14</sup> and Asp<sup>99</sup> are believed to participate in hydrogen bonding to the O-3 carbonyl of the substrate steroid and stabilize reaction intermediates. Tyr<sup>14</sup> is also believed to participate in a low barrier hydrogen bond with the 3-position oxygen of the steroid, thereby facilitating the abstraction of the β-hydrogen at the 4-position. There are two proposed models of this hydrogen bonding. In <scene name='User:Laura_M._Haynes/Sandbox_1/Model_1_1qjg/1'>Model 1</scene>, Tyr<sup>14</sup> and Asp<sup>99</sup> are both bound to the 3-position oxygen, whereas, in <scene name='User:Laura_M._Haynes/Sandbox_1/Model_2_1qjg/1'>Model 2</scene>, they form a hydrogen bonding network. These are <scene name='User:Laura_M._Haynes/Sandbox_1/1qjg/1'>shown here</scene> as a single monomer in complex with the intermediate analog [http://en.wikipedia.org/wiki/Equilenin equilenin].<ref name="Cho2">PMID:10551849</ref> | ||
| Line 75: | Line 76: | ||
Model 1 has become the generally accepted scheme through both crystallographic and mutanagenic/kinetic approaches. Mutation of Asp<sup>99</sup> to alanine and Tyr<sup>14</sup> to phenylalanine resulted in deleterious effects on kinetic parameters, which were additive in nature suggesting that Tyr<sup>14</sup> and Asp<sup>99</sup> participate equally in hydrogen binding with the oxygen atom. Additionally, the crystal structure of TI analog from ''Pseudomonas putida'' supports this conclusion given the <scene name='User:Laura_M._Haynes/Sandbox_1/Model_1_1qjg/1'>apparent hydrogen bonding distances</scene>.<ref name="Pollack" /> The general mechanism in the active site of KSI is outlined below. | Model 1 has become the generally accepted scheme through both crystallographic and mutanagenic/kinetic approaches. Mutation of Asp<sup>99</sup> to alanine and Tyr<sup>14</sup> to phenylalanine resulted in deleterious effects on kinetic parameters, which were additive in nature suggesting that Tyr<sup>14</sup> and Asp<sup>99</sup> participate equally in hydrogen binding with the oxygen atom. Additionally, the crystal structure of TI analog from ''Pseudomonas putida'' supports this conclusion given the <scene name='User:Laura_M._Haynes/Sandbox_1/Model_1_1qjg/1'>apparent hydrogen bonding distances</scene>.<ref name="Pollack" /> The general mechanism in the active site of KSI is outlined below. | ||
| - | [[Image:H_bond.jpg| | + | [[Image:H_bond.jpg|left|450px|thumb]] |
===Low Barrier Hydrogen Bond=== | ===Low Barrier Hydrogen Bond=== | ||
| Line 106: | Line 107: | ||
# Superfamily - NTF2-like | # Superfamily - NTF2-like | ||
# Family - Ketosteroid Isomserase-like | # Family - Ketosteroid Isomserase-like | ||
| - | # Domian - Δ5-3-ketosteroid isomerase | + | # Domian - Δ5-3-ketosteroid isomerase |
| + | </StructureSection> | ||
| + | __NOTOC__ | ||
| + | |||
| + | ==3D structures of ketosteroid isomerase== | ||
| + | |||
| + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| + | |||
| + | *Ketosteroid isomerase | ||
| + | |||
| + | **[[1isk]] – CtKSI – ''Comamonas testosteroni'' - NMR<BR /> | ||
| + | **[[8cho]] – CtKSI<BR /> | ||
| + | **[[1ocv]], [[3nxj]], [[3mhe]], [[3mki]], [[3myt]], [[3nm2]], [[3t8u]], [[3unl]], [[4l7k]], [[4k1u]], [[4k1v]], [[5dre]] – CtKSI (mutant) <BR /> | ||
| + | **[[1opy]], [[3vsy]], [[6u1z]], [[6ucw]] – PpKSI – ''Pseudomonas putida''<BR /> | ||
| + | **[[1c7h]], [[1dmm]], [[1dmn]], [[1dmq]], [[1e97]], [[1ea2]], [[1k41]], [[1vzz]], [[1w01]], [[1w02]], [[1w6y]], [[1w00]], [[3sed]], [[3ox9]], [[3oxa]], [[3t8n]], [[3rgr]], [[5d81]], [[5d82]], [[5d83]], [[6f4y]], [[6f50]], [[6f53]], [[6f54]], [[6uad]], [[6uae]], [[7rxf]], [[7rxk]] – PpKSI (mutant) <BR /> | ||
| + | **[[5z3r]] – KSI – ''Mycobacterium neoaurum'' <BR /> | ||
| + | **[[6p3l]] – MhKSI – ''Mycobacterium hassiacum'' <BR /> | ||
| + | **[[7epn]] – MsKSI – ''Mycobacterium smegmatis'' <BR /> | ||
| + | |||
| + | *Ketosteroid isomerase complexes | ||
| - | + | **[[1buq]] – CtKSI (mutant) + steroid - NMR<BR /> | |
| - | *[[ | + | **[[1qjg]], [[1ogz]], [[3m8c]], [[3ov4]], [[5ugi]] – CtKSI (mutant) + equilenin <BR /> |
| - | *[[ | + | **[[3nbr]], [[3nhx]], [[3nuv]] – CtKSI (mutant) + androgen derivative <BR /> |
| - | *[[ | + | **[[1qjg]] – PtKSI (mutant) + equilenin – ''Pseudomonas testosteroni''<BR /> |
| - | *[[1qjg]] | + | **[[1e3r]] – PpKSI (mutant) + androgen derivative <BR /> |
| - | *[[ | + | **[[1oh0]] – PpKSI + equilenin <BR /> |
| - | *[[ | + | **[[1gs3]], [[1ogx]], [[1cqs]], [[1oho]], [[3fzw]], [[3ipt]], [[3ows]], [[3owu]], [[3owy]], [[5ai1]], [[5g2g]], [[5kp1]], [[5kp3]], [[5kp4]] – PpKSI (mutant) + equilenin <BR /> |
| - | *[[ | + | **[[6tzd]], [[6ubq]], [[6ucy]] – PpKSI + androstenedione <BR /> |
| - | + | **[[6ufs]] – PpKSI + dihydronandrolone <BR /> | |
| - | + | **[[6u4i]], [[6ucn]] – PpKSI + equilenin <BR /> | |
| - | + | **[[1e3v]], [[4cdl]] – PpKSI + inhibitor <BR /> | |
| - | + | **[[2pzv]], [[2inx]], [[3cpo]], [[3vgn]], [[6c17]], [[6c1j]], [[6c1x]] – PpKSI (mutant) + phenol derivative<BR /> | |
| - | + | **[[7ry4]] – PpKSI (mutant) + transition state analog<BR /> | |
| - | + | **[[6p44]] – MhKSI (mutant) + phenol derivative<BR /> | |
| - | *[[ | + | **[[1ohp]] – CtKSI (mutant) + estrogen derivative <BR /> |
| - | + | **[[1ohs]] – CtKSI (mutant) + androgen derivative <BR /> | |
| - | + | **[[7epo]] – MsKSI + benzoxazole derivative <BR /> | |
| - | *[[ | + | }} |
| - | *[[ | + | |
| - | *[[ | + | |
| - | + | ||
| - | *[[2pzv]] | + | |
| - | + | ||
| - | *[[ | + | |
| - | *[[ | + | |
| - | *[[ | + | |
| - | *[[ | + | |
| - | *[[ | + | |
==References== | ==References== | ||
<references /> | <references /> | ||
| + | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
3D structures of ketosteroid isomerase
Updated on 13-June-2023
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 Pollack RM. Enzymatic mechanisms for catalysis of enolization: ketosteroid isomerase. Bioorg Chem. 2004 Oct;32(5):341-53. PMID:15381400 doi:10.1016/j.bioorg.2004.06.005
- ↑ 2.0 2.1 TALALAY P, WANG VS. Enzymic isomerization of delta5-3-ketosteroids. Biochim Biophys Acta. 1955 Oct;18(2):300-1. PMID:13276386
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 Ha NC, Choi G, Choi KY, Oh BH. Structure and enzymology of Delta5-3-ketosteroid isomerase. Curr Opin Struct Biol. 2001 Dec;11(6):674-8. PMID:11751047
- ↑ Ha NC, Choi G, Choi KY, Oh BH. Structure and enzymology of Delta5-3-ketosteroid isomerase. Curr Opin Struct Biol. 2001 Dec;11(6):674-8. PMID:11751047
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 Wu ZR, Ebrahimian S, Zawrotny ME, Thornburg LD, Perez-Alvarado GC, Brothers P, Pollack RM, Summers MF. Solution structure of 3-oxo-delta5-steroid isomerase. Science. 1997 Apr 18;276(5311):415-8. PMID:9103200
- ↑ 6.0 6.1 6.2 Murzin AG. How far divergent evolution goes in proteins. Curr Opin Struct Biol. 1998 Jun;8(3):380-7. PMID:9666335
- ↑ 7.0 7.1 Cleland WW, Frey PA, Gerlt JA. The low barrier hydrogen bond in enzymatic catalysis. J Biol Chem. 1998 Oct 2;273(40):25529-32. PMID:9748211
- ↑ 8.0 8.1 8.2 Kraut DA, Sigala PA, Pybus B, Liu CW, Ringe D, Petsko GA, Herschlag D. Testing electrostatic complementarity in enzyme catalysis: hydrogen bonding in the ketosteroid isomerase oxyanion hole. PLoS Biol. 2006 Apr;4(4):e99. Epub 2006 Mar 28. PMID:16602823 doi:10.1371/journal.pbio.0040099
- ↑ Cho HS, Choi G, Choi KY, Oh BH. Crystal structure and enzyme mechanism of Delta 5-3-ketosteroid isomerase from Pseudomonas testosteroni. Biochemistry. 1998 Jun 9;37(23):8325-30. PMID:9622484 doi:10.1021/bi9801614
- ↑ 10.0 10.1 Aurora R, Rose GD. Helix capping. Protein Sci. 1998 Jan;7(1):21-38. PMID:9514257 doi:10.1002/pro.5560070103
- ↑ 11.0 11.1 Kim DH, Jang DS, Nam GH, Choi KY. Folding mechanism of ketosteroid isomerase from Comamonas testosteroni. Biochemistry. 2001 Apr 24;40(16):5011-7. PMID:11305917
- ↑ Massiah MA, Abeygunawardana C, Gittis AG, Mildvan AS. Solution structure of Delta 5-3-ketosteroid isomerase complexed with the steroid 19-nortestosterone hemisuccinate. Biochemistry. 1998 Oct 20;37(42):14701-12. PMID:9778345 doi:10.1021/bi981447b
- ↑ Kim SW, Cha SS, Cho HS, Kim JS, Ha NC, Cho MJ, Joo S, Kim KK, Choi KY, Oh BH. High-resolution crystal structures of delta5-3-ketosteroid isomerase with and without a reaction intermediate analogue. Biochemistry. 1997 Nov 18;36(46):14030-6. PMID:9369474 doi:10.1021/bi971546+
- ↑ Sigala PA, Kraut DA, Caaveiro JM, Pybus B, Ruben EA, Ringe D, Petsko GA, Herschlag D. Testing geometrical discrimination within an enzyme active site: constrained hydrogen bonding in the ketosteroid isomerase oxyanion hole. J Am Chem Soc. 2008 Oct 15;130(41):13696-708. Epub 2008 Sep 23. PMID:18808119 doi:10.1021/ja803928m
- ↑ 15.0 15.1 Nam GH, Cha SS, Yun YS, Oh YH, Hong BH, Lee HS, Choi KY. The conserved cis-Pro39 residue plays a crucial role in the proper positioning of the catalytic base Asp38 in ketosteroid isomerase from Comamonas testosteroni. Biochem J. 2003 Oct 15;375(Pt 2):297-305. PMID:12852789 doi:10.1042/BJ20030263
- ↑ Cho HS, Ha NC, Choi G, Kim HJ, Lee D, Oh KS, Kim KS, Lee W, Choi KY, Oh BH. Crystal structure of delta(5)-3-ketosteroid isomerase from Pseudomonas testosteroni in complex with equilenin settles the correct hydrogen bonding scheme for transition state stabilization. J Biol Chem. 1999 Nov 12;274(46):32863-8. PMID:10551849
- ↑ 17.0 17.1 Zhao Q, Abeygunawardana C, Talalay P, Mildvan AS. NMR evidence for the participation of a low-barrier hydrogen bond in the mechanism of delta 5-3-ketosteroid isomerase. Proc Natl Acad Sci U S A. 1996 Aug 6;93(16):8220-4. PMID:8710850
- ↑ Zhao Q, Abeygunawardana C, Talalay P, Mildvan AS. NMR evidence for the participation of a low-barrier hydrogen bond in the mechanism of delta 5-3-ketosteroid isomerase. Proc Natl Acad Sci U S A. 1996 Aug 6;93(16):8220-4. PMID:8710850
- ↑ 19.0 19.1 Kraut DA, Sigala PA, Fenn TD, Herschlag D. Dissecting the paradoxical effects of hydrogen bond mutations in the ketosteroid isomerase oxyanion hole. Proc Natl Acad Sci U S A. 2010 Feb 2;107(5):1960-5. Epub 2010 Jan 11. PMID:20080683
- ↑ Sigala PA, Caaveiro JM, Ringe D, Petsko GA, Herschlag D. Hydrogen bond coupling in the ketosteroid isomerase active site. Biochemistry. 2009 Jul 28;48(29):6932-9. PMID:19469568 doi:10.1021/bi900713j
- ↑ 21.0 21.1 Cherney MM, Garen CR, James MN. Crystal structure of Mycobacterium tuberculosis Rv0760c at 1.50 A resolution, a structural homolog of Delta(5)-3-ketosteroid isomerase. Biochim Biophys Acta. 2008 Nov;1784(11):1625-32. Epub 2008 Jun 6. PMID:18589008 doi:10.1016/j.bbapap.2008.05.012
Proteopedia Page Contributors and Editors (what is this?)
Laura M. Haynes, Michal Harel, Joel L. Sussman, Alexander Berchansky