CAMP Dependent Protein Kinase, Catalytic Subunit
From Proteopedia
(Difference between revisions)
| (7 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='1j3h' size='350' side='right' scene='' caption='Mouse CAMP-dependent protein kinase cdatalytic subunit with phosphothreonine, phosphoserine and MPD [[1j3h]]'> | |
==Introduction== | ==Introduction== | ||
| Line 8: | Line 8: | ||
==Activation and Interaction with Regulatory Subunit== | ==Activation and Interaction with Regulatory Subunit== | ||
| - | [[Image:PKA1.svg.png]] [[Image:200px-CAMP.svg.png]] | ||
| - | |||
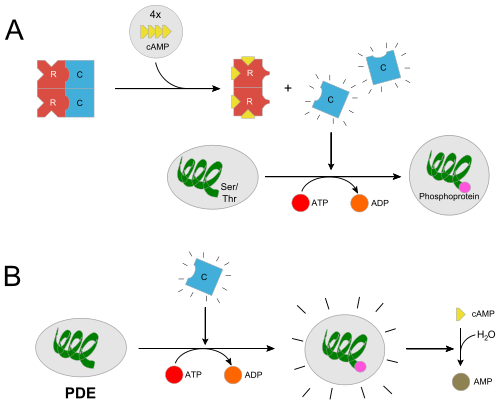
In the inactive state, the catalytic subunit of PKA exists as a heterotetramer with two regulatory subunits and two catalytic subunits. Regulatory subunits often interact with A Kinase Anchoring proteins that serve to localize a population of PKA in a certain cellular environment, priming a particular response. Upon, cAMP binding to the regulatory domain of PKA (two molecules of cAMP per regulatory subunit) the catalytic subunit is released from the holoenzyme complex and is free to diffuse and exhibit its catalytic activity. Two of the most important residues for this docking interaction are <scene name='CAMP_Dependent_Protein_Kinase,_Catalytic_Subunit/Tyr247/1'>Trp196</scene> and <scene name='CAMP_Dependent_Protein_Kinase,_Catalytic_Subunit/Tyr247/3'>Tyr247</scene> which both sit in the nucleotide binding region (Phosphate Binding Cassette) of the regulatory subunit. In essence, when a cAMP molecule binds to these trp and tyr binding sites, the docking interaction is ablated. This <scene name='CAMP_Dependent_Protein_Kinase,_Catalytic_Subunit/Hydro-contacts/1'>scene</scene> shows a different representation. <ref>PMID: 17889648</ref> <ref>PMID: 3356685 | In the inactive state, the catalytic subunit of PKA exists as a heterotetramer with two regulatory subunits and two catalytic subunits. Regulatory subunits often interact with A Kinase Anchoring proteins that serve to localize a population of PKA in a certain cellular environment, priming a particular response. Upon, cAMP binding to the regulatory domain of PKA (two molecules of cAMP per regulatory subunit) the catalytic subunit is released from the holoenzyme complex and is free to diffuse and exhibit its catalytic activity. Two of the most important residues for this docking interaction are <scene name='CAMP_Dependent_Protein_Kinase,_Catalytic_Subunit/Tyr247/1'>Trp196</scene> and <scene name='CAMP_Dependent_Protein_Kinase,_Catalytic_Subunit/Tyr247/3'>Tyr247</scene> which both sit in the nucleotide binding region (Phosphate Binding Cassette) of the regulatory subunit. In essence, when a cAMP molecule binds to these trp and tyr binding sites, the docking interaction is ablated. This <scene name='CAMP_Dependent_Protein_Kinase,_Catalytic_Subunit/Hydro-contacts/1'>scene</scene> shows a different representation. <ref>PMID: 17889648</ref> <ref>PMID: 3356685 | ||
</ref> | </ref> | ||
| + | |||
| + | |||
| + | [[Image:PKA1.svg.png]] [[Image:200px-CAMP.svg.png]] | ||
==Binding to ATP== | ==Binding to ATP== | ||
| - | Typical kinases are characterized by 3 highly conserved glycine residues located near the junction between the small and large kinase subunits. The | + | Typical kinases are characterized by 3 highly conserved glycine residues located near the junction between the small and large kinase subunits. The loop region that these occur in is referred to as the <scene name='CAMP_Dependent_Protein_Kinase,_Catalytic_Subunit/Gly-atp-close/1'>glycine-rich loop</scene>. Mutation of any of these residues results in dramatic reduction of kinase affinity for ATP. The structural motif that the glycine rich loop is located in is a beta-strand, turn, beta-strand motif. These residues are also reported to have some role in phosphoryl transfer. <ref>PMID: 9202006</ref> |
| + | |||
| + | ==Interaction with Substrate== | ||
| + | The larger C-terminal lobe of the kinase is responsible for substrate recognition. Several of the c-term lobes alpha helices are important in determining substrate recognition. The substrate mimic inhibitor <scene name='CAMP_Dependent_Protein_Kinase,_Catalytic_Subunit/Pki/1'>PKI</scene> demonstrates this nicely. The PKI inhibitor is a heat shock protein. It sits firmly within the catalytic cleft of the enzyme. <ref>PMID: 8443157</ref> | ||
| + | </StructureSection> | ||
| + | ==3D structures of CAMP-dependent protein kinase== | ||
| + | |||
| + | [[CAMP-dependent protein kinase]] | ||
| + | |||
==References== | ==References== | ||
<references /> | <references /> | ||
Current revision
| |||||||||||
3D structures of CAMP-dependent protein kinase
References
- ↑ Taylor SS, Radzio-Andzelm E, Knighton DR, Ten Eyck LF, Sowadski JM, Herberg FW, Yonemoto W, Zheng J. Crystal structures of the catalytic subunit of cAMP-dependent protein kinase reveal general features of the protein kinase family. Receptor. 1993 Fall;3(3):165-72. PMID:8167567
- ↑ Yonemoto W, McGlone ML, Taylor SS. N-myristylation of the catalytic subunit of cAMP-dependent protein kinase conveys structural stability. J Biol Chem. 1993 Feb 5;268(4):2348-52. PMID:8428909
- ↑ Kim C, Cheng CY, Saldanha SA, Taylor SS. PKA-I holoenzyme structure reveals a mechanism for cAMP-dependent activation. Cell. 2007 Sep 21;130(6):1032-43. PMID:17889648 doi:10.1016/j.cell.2007.07.018
- ↑ First EA, Bubis J, Taylor SS. Subunit interaction sites between the regulatory and catalytic subunits of cAMP-dependent protein kinase. Identification of a specific interchain disulfide bond. J Biol Chem. 1988 Apr 15;263(11):5176-82. PMID:3356685
- ↑ Hemmer W, McGlone M, Tsigelny I, Taylor SS. Role of the glycine triad in the ATP-binding site of cAMP-dependent protein kinase. J Biol Chem. 1997 Jul 4;272(27):16946-54. PMID:9202006
- ↑ Zheng J, Knighton DR, ten Eyck LF, Karlsson R, Xuong N, Taylor SS, Sowadski JM. Crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MgATP and peptide inhibitor. Biochemistry. 1993 Mar 9;32(9):2154-61. PMID:8443157