We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
A-ATP Synthase
From Proteopedia
(Difference between revisions)
| (32 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | [[Image: | + | <StructureSection load=1e79 size='450' side='right' caption='Bovine ATP synthase chains α (grey, green, pink), β (yellow, magenta, cyan), γ (gold), δ (red), ε (wheat) complex with ATP (stick model), ADP (stick model), glycerol, dicyclohexylurea, sulfate and Mg+2 ions (green) (PDB code [[1e79]])' scene=''> |
| + | [[Image:F_ATPsynthase.gif | thumb|left]] | ||
| + | __NOTOC__ | ||
| + | ==Structure== | ||
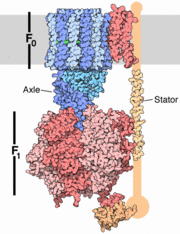
| + | The structure on the right shows the F1 motor and the axle that connects the two. ATP synthesis is composed of two rotary motors, each powered by a different fuel. The motor at the top, termed F0, an electric motor. It is embedded in a membrane (shown schematically as a gray stripe here), and is powered by the flow of hydrogen ions across the membrane. As the protons flow through the motor, they turn a circular rotor . This rotor is connected to the second motor, termed F1. The F1 motor is a chemical motor, powered by ATP. The two motors are connected together by a stator, shown on the right, so that when F0 turns, F1 turns too. [http://en.wikipedia.org/wiki/Atp_synthase A-ATP synthase] is very similar to F ATP Synthase and is composed of two parts '''A1''' and '''A0''' which are composed of at least nine subunits '''A3B3C:D:E:F:H2:a:cx''' that function as a pair of rotary motors connected by central and peripheral stalk(s) <ref name= Muller> PMID: 16645313</ref>. The '''A0''' domain is the hydrophobic membrane embedded ion-translocating sector that uses the H+ gradient to power ATP synthase in domain '''A1'''. '''A1''' is catalytic and water soluble containing '''A''' and '''B''' subunits. These subunits are comparable to F-ATP synthase [http://en.wikipedia.org/wiki/ATP_synthase_alpha/beta_subunits ATP synthase alpha/beta subunits]. The '''A''' subunit of '''A1''' is catalytic and the '''B''' subunit is regulatory, with a substrate-binding site on each. [http://www.youtube.com/watch?v=KU-B7G6anqw&feature=related] | ||
| + | |||
| + | ==<scene name='A-ATP_Synthase/Rotary_stalk/1'>F1 ATP Synthase Rotary Mechanism</scene>== | ||
| + | |||
| + | The central stalk in ATP synthase, made of gamma, delta and epsilon subunits in the mitochondrial enzyme, is the key rotary element in the enzyme's catalytic mechanism. The <scene name='A-ATP_Synthase/Rotary_stalk/12'>Gamma subunit</scene> penetrates the catalytic (alpha beta)(3) domain and protrudes beneath it, interacting with a ring of c subunits in the membrane that drives rotation of the stalk during ATP synthesis. In ATP synthase, the central stalk interacts with the c-ring and couples the transmembrane proton motive force to catalysis in the (<scene name='A-ATP_Synthase/Rotary_stalk/8'>alpha, beta</scene> (3) domain. | ||
| + | |||
| + | When operating as a generator, it uses the power of rotational motion to build ATP, or when operating as a motor, it breaks down ATP to spin the axle the opposite direction. The synthesis of ATP requires <scene name='A-ATP_Synthase/Rotary_stalk/4'>several steps</scene> , including the binding of ADP and phosphate, the formation of the new phosphate-phosphate bond, and release of ATP. As the axle turns, it forces the motor into three different conformations that assist these difficult steps. The beta subunits have a structural role, holding everything in place. The alpha subunits are the ATP generating parts. <ref name= Gibbons> PMID:11062563</ref>There are three catalytic nucleotide binding sites and three corresponding states induced by the central stalks rotation. In the Alternating catalytic model, the binding sites go through three different states. In the Open State ATP is released and ADP and Pi enters the active site. In the Loose state ADP and Pi are bound and the active site closes up around the molecules.The Tight State forces molecules together, catalyzing the formation of the phosphate bond. | ||
| + | |||
==Introduction== | ==Introduction== | ||
| Line 6: | Line 17: | ||
<ref name= Gonzales> PMID:9672687</ref> | <ref name= Gonzales> PMID:9672687</ref> | ||
The specific enzymatic process in A-ATP synthase reveals novel, exceptional subunit composition and coupling stoichiometries that may reflect the differences in energy-conserving mechanisms as well as adaptation to temperatures at or above 100 degrees C. Because some [http://en.wikipedia.org/wiki/Archaea archaea] are rooted close to the origin in the tree of life, these unusual | The specific enzymatic process in A-ATP synthase reveals novel, exceptional subunit composition and coupling stoichiometries that may reflect the differences in energy-conserving mechanisms as well as adaptation to temperatures at or above 100 degrees C. Because some [http://en.wikipedia.org/wiki/Archaea archaea] are rooted close to the origin in the tree of life, these unusual | ||
| - | mechanisms are considered to have developed very early in the history of life and, therefore, may represent the first energy-conserving mechanisms. <ref name= Muller> PMID: 16645313</ref> | + | mechanisms are considered to have developed very early in the history of life and, therefore, may represent the first energy-conserving mechanisms. <ref name= Muller> PMID: 16645313</ref> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | ==Structure of A-ATP synthase catalytic subunit A== | ||
| - | = | + | Five steps inside the catalytic A-subunit are critical for catalysis. Substrate entrance, phosphate and nucleotide binding, transition-state formation, ATP formation, and product release. The [http://en.wikipedia.org/wiki/Vanadate vanadate] bound model mimics the transition state. [http://en.wikipedia.org/wiki/Orthovanadate Orthovandate] is a useful transition state analog because it can adapt both tetragonal and trigonal bipyramidal coordination geometry. The '''Avi''' structure can be compared to the '''As''' sulfate bound structure and the '''Apnp''' AMP-PNP bound structure. '''As''' is analogous to the phosphate binding (substrate) structure and '''Apnp''' is analogous to the ATP binding (product) structure<ref name= Manimekalai> PMID:21396943</ref>. |
| - | Five steps inside the catalytic A-subunit are critical for catalysis. Substrate entrance, phosphate and nucleotide binding, transition-state formation, ATP formation, and product release. The [http://en.wikipedia.org/wiki/Vanadate vanadate] bound model mimics the transition state. [http://en.wikipedia.org/wiki/Orthovanadate Orthovandate] is a useful transition state analog because it can adapt both tetragonal and trigonal bipyramidal coordination geometry. The '''Avi''' structure can be compared to the '''As''' sulfate bound structure and the '''Apnp''' AMP-PNP bound structure. '''As''' is analogous to the phosphate binding (substrate) structure and '''Apnp''' is analogous to the ATP binding (product) structure<ref name= Manimekalai?> PMID:21396943</ref>. The movement of specific residues to stabilize the transition state is demonstrated by comparing the deviations between the three structures. Although not at bonding distances the residues P233 G234 L417 stabilize the first <scene name='A-ATP_Synthase/Vandates-2/3'>vandate</scene> in the transition state with weak nonpoalr interactions, and residues K240 and T241 stabilize with polar interactions. | ||
| - | + | Within the catalytic '''A''' subunit there are <scene name='A-ATP_Synthase/3p20_main_structure/4'>four domains,</scene> the '''N-terminal domain''' residues 1-79, 110-116, 189-199, the '''non-homologous region'''(which region makes this subunit considerably larger than its homologs) residues 117-188, '''the nucleotide binding alpha-beta domain''' residues 80-99, 200-437, and '''C-terminal alpha helical bundle''' residues 438-588 domains. There are 16 helices and 27 strands. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ==P-Loop== | |
| + | The <scene name='A-ATP_Synthase/3p20_main_structure/8'>P-Loop</scene>is the eight residue consensus sequence of amino acid residues 234-241 '''G'''PFGS'''GKT''' . The P-loop or phosphate binding loop is conserved only within the A subunits and is a <scene name='A-ATP_Synthase/3p20_main_structure/7'>glycine rich</scene> loop preceded by a beta sheet and followed by an alpha helix. | ||
| + | |||
| + | <scene name='A-ATP_Synthase/Vanadate_1_interactions/2'>Vanadate one</scene> occupies the ADP site. Although not at bonding distances the residues P233 G234 L417 stabilize the first vanadate in the transition state with weak nonpolar interactions. Residues K240 and T241 stabilize with polar interactions. | ||
| + | Residue <scene name='A-ATP_Synthase/238/6'>S238</scene> is a polar serine molecule that interacts with the nucleotides via a hydrogen bond during catalysis. The distance between residue S238 is longest in '''As''', shortest in '''Avi''' and intermediate in '''Apnp''' . In '''As''' a water molecule bridges the gap, which is removed in '''Avi'''. Dehydration of the transition state active site is reversed when ATP forms. In '''Apnp''' the water molecule interacts with the y-phosphate of ATP. | ||
| + | ==Comparisons to other known structures== | ||
| + | This P-loop has an arched conformation unique to A-ATP Synthase, indicating that the mode of nucleotide binding and the catalytic mechanism is different from that of other syntheses. <ref name= Priya> PMID: 21925186</ref> | ||
| + | The P-loop is unique in archea because it is stable compared to other similar structures. For example, in A-ATP Synthases <scene name='A-ATP_Synthase/P_loop_3p20/12'>F236</scene> is involved in P-Loop stabilization. | ||
| + | Another reason why the P-Loop is so stable in A-ATP synthase is because of residue S238. It is significant in the catalytic process in moving towards the y-phosphate of ATP during catalysis, and does this with a hydrogen bond, not a conformational change in the P-loop. In "'F-ATP Synthase"' the homolog to S238 is the non polar A158. Since A158 cannot form hydrogen bonds to interact with the substrate (like s238), the P-loop undergoes a conformational change.<ref name= Manimekalai> PMID:21396943</ref> | ||
</StructureSection> | </StructureSection> | ||
| - | {{STRUCTURE_3p20| PDB=3p20 | SIZE=400| SCENE= |right|CAPTION=Transition state, [[3p20]] }} | ||
| - | {{TOC limit|limit=2}} | ||
==References== | ==References== | ||
{{Reflist}} | {{Reflist}} | ||
Current revision
| |||||||||||
References
- ↑ 1.0 1.1 Muller V, Lemker T, Lingl A, Weidner C, Coskun U, Gruber G. Bioenergetics of archaea: ATP synthesis under harsh environmental conditions. J Mol Microbiol Biotechnol. 2005;10(2-4):167-80. PMID:16645313 doi:10.1159/000091563
- ↑ Gibbons C, Montgomery MG, Leslie AG, Walker JE. The structure of the central stalk in bovine F(1)-ATPase at 2.4 A resolution. Nat Struct Biol. 2000 Nov;7(11):1055-61. PMID:11062563 doi:10.1038/80981
- ↑ Schafer IB, Bailer SM, Duser MG, Borsch M, Bernal RA, Stock D, Gruber G. Crystal structure of the archaeal A1Ao ATP synthase subunit B from Methanosarcina mazei Go1: Implications of nucleotide-binding differences in the major A1Ao subunits A and B. J Mol Biol. 2006 May 5;358(3):725-40. Epub 2006 Mar 10. PMID:16563431 doi:http://dx.doi.org/10.1016/j.jmb.2006.02.057
- ↑ Gonzalez JM, Masuchi Y, Robb FT, Ammerman JW, Maeder DL, Yanagibayashi M, Tamaoka J, Kato C. Pyrococcus horikoshii sp. nov., a hyperthermophilic archaeon isolated from a hydrothermal vent at the Okinawa Trough. Extremophiles. 1998 May;2(2):123-30. PMID:9672687
- ↑ 5.0 5.1 Manimekalai MS, Kumar A, Jeyakanthan J, Gruber G. The Transition-Like State and P(i) Entrance into the Catalytic A Subunit of the Biological Engine A-ATP Synthase. J Mol Biol. 2011 Mar 16. PMID:21396943 doi:10.1016/j.jmb.2011.03.010
- ↑ Priya R, Kumar A, Manimekalai MS, Gruber G. Conserved Glycine Residues in the P-Loop of ATP Synthases Form a Doorframe for Nucleotide Entrance. J Mol Biol. 2011 Sep 8. PMID:21925186 doi:10.1016/j.jmb.2011.08.045
Proteopedia Page Contributors and Editors (what is this?)
Kaitlin Chase MacCulloch, Michal Harel, Alexander Berchansky