Sandbox Reserved 383
From Proteopedia
(→Ligands) |
|||
| (62 intermediate revisions not shown.) | |||
| Line 4: | Line 4: | ||
| - | ''' | + | ==Human Dopamine D3 Receptor== |
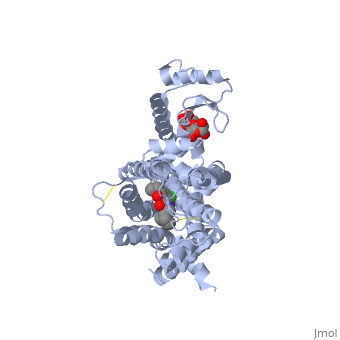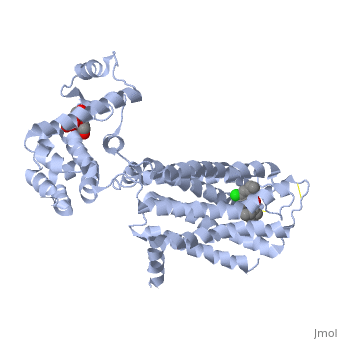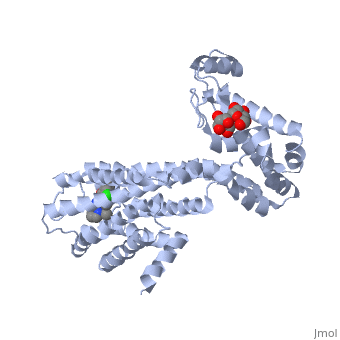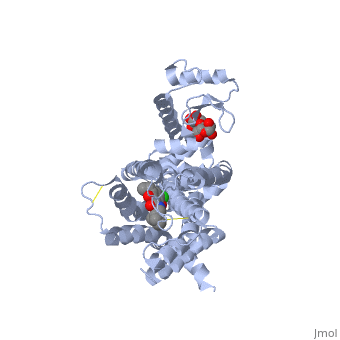
| + | <Structure load='3PBL' size='500' frame='true' align='right' caption='Structure of the human dopamine D3 receptor in complex with eticlopride' scene='Insert optional scene name here' /> | ||
| + | ==Introduction== | ||
| + | Dopamine receptors are a class of metabotropic G protein-coupled receptors that are important in the central nervous system. Dopamine receptors are involved in many neurological processes that comprise motivation, pleasure, cognition, memory, learning, and fine motor skills. There are five subtype dopamine receptors, D1, D2, D3, D4, and D5. The D3 receptor is a part of the D2-like family.<ref>PMID:15148138</ref> | ||
| - | == ' | + | ==Function== |
| + | Human dopamine D3 receptor is a protein that is encoded by the dopamine receptor gene (DRD3).<ref>PMID: 1916765</ref> The DRD3 gene codes for the D3 dopamine receptor that inhibits adenylyl cyclase through inhibitory G-proteins. G-protein coupled receptors are a family of transmembrane proteins that transmit chemical signals from outside the cell to cause changes inside of the cell. Adenylate cyclase is part of the G-protein receptor's signaling and catalyze the conversion of ATP to cyclic AMP (cAMP).<ref>Reece, Jane; Campbell, Neil (2002). Biology. San Francisco: Benjamin Cummings. ISBN 0-8053-6624-5.</ref> The D3 receptor is located in the brain, suggesting that it plays a role in cognitive and emotional functions.<ref>National Center for Biotechnology Information, U.S. National Library of Medicine. DRD3 dopamine receptor D3 [Homo sapiens]. 19 November 2011.</ref> The human dopamine D3 receptor is membrane-bound and scattered in the cytoplasm. Receptor stimulation causes internalization of the receptors at the perinuclear areas. This is followed by the spreading of the receptors to the membrane. DRD3 is also contained in lipid rafts of renal proximal tubule cells.<ref>PMID: 19520868</ref> | ||
| - | + | ==Structure== | |
| + | Human dopamine D3 receptor is 64% helical and 1% beta sheet. The protein is composed of 20 helices and 3 beta sheet strands. The helices are made up of 312 residues, and the beta sheets are made up of 9 residues. The entire protein consists of 481 residues.<ref name="structure">PMID: 21097933</ref> Dopamine D3 receptors are greatly expressed in the Islands of Calleja, a group of neural granule cells located within the ventral striatum in the brains of most animals, which is part of the limbic system. It is also found in the nucleus accumbens, a collection of neurons, and forms the main part of the ventral striatum.<ref>PMID:9473588</ref> | ||
| - | == ''' | + | ==Ligands== |
| + | Many non-selective prescription drugs bind to the D3 receptor. The binding of drugs either increases or inhibits the production of dopamine D3 receptors, which helps to diminish complications that are caused by certain diseases. Some agonists, agents that stimulate dopamine receptors, include: | ||
| + | *Amphetamine<ref>Jones S, Kornblum JL, Kauer JA (August 2000). "Amphetamine blocks long-term synaptic depression in the ventral tegmental area". J. Neurosci. 20 (15): 5575–80. PMID 10908593. http://www.jneurosci.org/cgi/pmidlookup?view=long&pmid=10908593.</ref> | ||
| + | *Methamphetamine<ref>Cruickshank, CC.; Dyer, KR. (Jul 2009). "A review of the clinical pharmacology of methamphetamine.". Addiction 104 (7): 1085–99. doi:10.1111/j.1360-0443.2009.02564.x. PMID 19426289.</ref> | ||
| + | Some antagonists, agents that inhibit dopamine receptors, include: | ||
| + | *Clebopride<ref>Cuena Boy R, Maciá Martínez MA (1998). "[Extrapyramidal toxicity caused by metoclopramide and clebopride: study of voluntary notifications of adverse effects to the Spanish Drug Surveillance System]" (in Spanish). Atencion Primaria 21 (5): 289–95. PMID 9608114. Free full text</ref> | ||
| + | *Nafadotride<ref>Pilla M, Perachon S, Sautel F, Garrido F, Mann A, Wermuth CG, Schwartz JC, Everitt BJ, Sokoloff P. Selective inhibition of cocaine-seeking behaviour by a partial dopamine D3 agonist. Nature. 1999;400:371–375.</ref> | ||
| + | Two ligands that are associated with the dopamine D3 receptor are 3-chloro-5-ethyl-N{[(2S)-1-ethylpyrrolidin-2-yl]methyl}-6-hydroxyl-2-methyloxybenzamide (<scene name='Sandbox_Reserved_383/Etq/1'>ETQ</scene>) and maltose (<scene name='Sandbox_Reserved_383/Mal/1'>MAL</scene>). ETQ binds to dopamine D3 receptor by Asp 110A and Phe346A. MAL binds by Asp 1020A, Glu 1022A, Glu1011A, and Leu1032A.<ref name="structure" /> | ||
| - | + | ==Diseases== | |
| + | Variations in the DRD3 gene is connected with essential tremor hereditary type 1 (ETM1).<ref name="disease">PMID: 16650084</ref> | ||
| + | ETM1 is the most common movement disorder involving postural tremor of the arms, head, legs, body core, voice, jaw, and other facial muscles. | ||
| + | This condition can be provoked by emotions, hunger, fatigue, and temperature extremes.<ref name="disease" /> | ||
| - | == | + | ==References== |
| - | + | <references /> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
| This Sandbox is Reserved from September 14, 2021, through May 31, 2022, for use in the class Introduction to Biochemistry taught by User:John Means at the University of Rio Grande, Rio Grande, OH, USA. This reservation includes 5 reserved sandboxes (Sandbox Reserved 1590 through Sandbox Reserved 1594). |
To get started:
More help: Help:Editing. For an example of a student Proteopedia page, please see Photosystem II, Tetanospasmin, or Guanine riboswitch. |
Contents |
Human Dopamine D3 Receptor
|
Introduction
Dopamine receptors are a class of metabotropic G protein-coupled receptors that are important in the central nervous system. Dopamine receptors are involved in many neurological processes that comprise motivation, pleasure, cognition, memory, learning, and fine motor skills. There are five subtype dopamine receptors, D1, D2, D3, D4, and D5. The D3 receptor is a part of the D2-like family.[1]
Function
Human dopamine D3 receptor is a protein that is encoded by the dopamine receptor gene (DRD3).[2] The DRD3 gene codes for the D3 dopamine receptor that inhibits adenylyl cyclase through inhibitory G-proteins. G-protein coupled receptors are a family of transmembrane proteins that transmit chemical signals from outside the cell to cause changes inside of the cell. Adenylate cyclase is part of the G-protein receptor's signaling and catalyze the conversion of ATP to cyclic AMP (cAMP).[3] The D3 receptor is located in the brain, suggesting that it plays a role in cognitive and emotional functions.[4] The human dopamine D3 receptor is membrane-bound and scattered in the cytoplasm. Receptor stimulation causes internalization of the receptors at the perinuclear areas. This is followed by the spreading of the receptors to the membrane. DRD3 is also contained in lipid rafts of renal proximal tubule cells.[5]
Structure
Human dopamine D3 receptor is 64% helical and 1% beta sheet. The protein is composed of 20 helices and 3 beta sheet strands. The helices are made up of 312 residues, and the beta sheets are made up of 9 residues. The entire protein consists of 481 residues.[6] Dopamine D3 receptors are greatly expressed in the Islands of Calleja, a group of neural granule cells located within the ventral striatum in the brains of most animals, which is part of the limbic system. It is also found in the nucleus accumbens, a collection of neurons, and forms the main part of the ventral striatum.[7]
Ligands
Many non-selective prescription drugs bind to the D3 receptor. The binding of drugs either increases or inhibits the production of dopamine D3 receptors, which helps to diminish complications that are caused by certain diseases. Some agonists, agents that stimulate dopamine receptors, include:
Some antagonists, agents that inhibit dopamine receptors, include:
Two ligands that are associated with the dopamine D3 receptor are 3-chloro-5-ethyl-N{[(2S)-1-ethylpyrrolidin-2-yl]methyl}-6-hydroxyl-2-methyloxybenzamide () and maltose (). ETQ binds to dopamine D3 receptor by Asp 110A and Phe346A. MAL binds by Asp 1020A, Glu 1022A, Glu1011A, and Leu1032A.[6]
Diseases
Variations in the DRD3 gene is connected with essential tremor hereditary type 1 (ETM1).[12]
ETM1 is the most common movement disorder involving postural tremor of the arms, head, legs, body core, voice, jaw, and other facial muscles. This condition can be provoked by emotions, hunger, fatigue, and temperature extremes.[12]
References
- ↑ Girault JA, Greengard P. The neurobiology of dopamine signaling. Arch Neurol. 2004 May;61(5):641-4. PMID:15148138 doi:10.1001/archneur.61.5.641
- ↑ Le Coniat M, Sokoloff P, Hillion J, Martres MP, Giros B, Pilon C, Schwartz JC, Berger R. Chromosomal localization of the human D3 dopamine receptor gene. Hum Genet. 1991 Sep;87(5):618-20. PMID:1916765
- ↑ Reece, Jane; Campbell, Neil (2002). Biology. San Francisco: Benjamin Cummings. ISBN 0-8053-6624-5.
- ↑ National Center for Biotechnology Information, U.S. National Library of Medicine. DRD3 dopamine receptor D3 [Homo sapiens]. 19 November 2011.
- ↑ Villar VA, Jones JE, Armando I, Palmes-Saloma C, Yu P, Pascua AM, Keever L, Arnaldo FB, Wang Z, Luo Y, Felder RA, Jose PA. G protein-coupled receptor kinase 4 (GRK4) regulates the phosphorylation and function of the dopamine D3 receptor. J Biol Chem. 2009 Aug 7;284(32):21425-34. Epub 2009 Jun 11. PMID:19520868 doi:10.1074/jbc.M109.003665
- ↑ 6.0 6.1 Chien EY, Liu W, Zhao Q, Katritch V, Han GW, Hanson MA, Shi L, Newman AH, Javitch JA, Cherezov V, Stevens RC. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010 Nov 19;330(6007):1091-5. PMID:21097933 doi:10.1126/science.1197410
- ↑ Suzuki M, Hurd YL, Sokoloff P, Schwartz JC, Sedvall G. D3 dopamine receptor mRNA is widely expressed in the human brain. Brain Res. 1998 Jan 1;779(1-2):58-74. PMID:9473588
- ↑ Jones S, Kornblum JL, Kauer JA (August 2000). "Amphetamine blocks long-term synaptic depression in the ventral tegmental area". J. Neurosci. 20 (15): 5575–80. PMID 10908593. http://www.jneurosci.org/cgi/pmidlookup?view=long&pmid=10908593.
- ↑ Cruickshank, CC.; Dyer, KR. (Jul 2009). "A review of the clinical pharmacology of methamphetamine.". Addiction 104 (7): 1085–99. doi:10.1111/j.1360-0443.2009.02564.x. PMID 19426289.
- ↑ Cuena Boy R, Maciá Martínez MA (1998). "[Extrapyramidal toxicity caused by metoclopramide and clebopride: study of voluntary notifications of adverse effects to the Spanish Drug Surveillance System]" (in Spanish). Atencion Primaria 21 (5): 289–95. PMID 9608114. Free full text
- ↑ Pilla M, Perachon S, Sautel F, Garrido F, Mann A, Wermuth CG, Schwartz JC, Everitt BJ, Sokoloff P. Selective inhibition of cocaine-seeking behaviour by a partial dopamine D3 agonist. Nature. 1999;400:371–375.
- ↑ 12.0 12.1 Lucotte G, Lagarde JP, Funalot B, Sokoloff P. Linkage with the Ser9Gly DRD3 polymorphism in essential tremor families. Clin Genet. 2006 May;69(5):437-40. PMID:16650084 doi:10.1111/j.1399-0004.2006.00600.x