Brittany deRonde/Sandbox 1
From Proteopedia
| (65 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | One of the [[CBI Molecules]] being studied in the [http://www.umass.edu/cbi/ University of Massachusetts Amherst Chemistry-Biology Interface Program] at UMass Amherst and on display at the [http://www.molecularplayground.org/ Molecular Playground]. | |
| - | <applet load='1ema' size='450' frame='true' align='right' scene='Green_Fluorescent_Protein/1ema_gfp_default/2' caption='Green fluorescent protein complex with peptide-derived chromophore ([[1ema]])' /> | ||
| - | '''HIV Tat''' is a [[bioluminescent]] polypeptide consisting of 238 residues isolated from the body of [[Aequorea victoria]] jellyfish.<ref name="PDBsum">[http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=1ema&template=main.html], Protein Database (PDBsum): 1ema. European Bioinformatics (EBI); 2009.</ref> GFP converts the blue chemiluminescent of [[aequorin]] in the jellyfish into green fluorescent light.<ref name="Yang">[http://www-bioc.rice.edu/Bioch/Phillips/Papers/gfpbio.html], Yang F, Moss LG, Phillips GN Jr. 1996. The molecular structure of green fluorescent protein. Biotechnology. 14: 1246-1251. DOI 10.1038/nbt1096-1246.</ref> It remains unclear why these jellyfish use fluorescence, why green is better than blue, or why they produce a separate protein for green fluorescence as opposed to simply mutating the present aequorin to shift its wavelength,<ref name="Tsien" /> but in the laboratory, GFP can be incorporated into a variety of biological systems in order to function as a marker protein. Since its discovery in 1962, GFP has become a significant contributor to the research of monitoring gene expression, localization, mobility, traffic, interactions between various membrane and cytoplasmic proteins, as well as many others.<ref name="Haldar">[http://www.springerlink.com/content/wvg513864266g77n/fulltext.pdf], Haldar S, Chattopadhyay A. 2009. The green journey. J Fluoresc. 19:1-2. DOI 10.1007/s10895-008-0455-6; biographical background on [http://en.wikipedia.org/wiki/Douglas_Prasher Douglas Prasher], [http://en.wikipedia.org/wiki/Martin_Chalfie Martin Chalfie] and [http://en.wikipedia.org/wiki/Roger_Tsien Roger Tsien].</ref> | ||
| - | == | + | == '''Introduction''' == |
| - | '' | + | '''HIV Tat''', or simply Tat, is a human immunodeficiency virus (HIV) gene that regulates transcription of HIV dsRNA.<ref>Sodroski ''et al. Science.'' '''1985'''. 227, 171-173. [http://www.jstor.org/stable/1695050] http://www.jstor.org/stable/1695050</ref> Tat, which stands for trans-activator of transcription, contains 86 amino acid residues in its sequence.<ref>Arya ''et al. Science'' '''1985'''. 229, 69-73.</ref><ref>Sodroski ''et al. Science.'' '''1985'''. 229, 74-77.</ref> <Structure load='1TIV' size='500' frame='true' align='right' caption='HIV Tat. Grey residues = polar groups, Pink residues= hydrophobic residues, and Blue residues= protein transduction domain' scene='Brittany_deRonde/Sandbox_1/Hiv_tat/2' /> It is released by HIV infected cells in order to enhance replication of the virus.<ref>Dayton ''et al. Cell.'' '''1986'''. 44, 941-947.</ref><ref>Fisher ''et al. Nature.'' '''1986'''. 320, 367-371.</ref> Nanomolar concentrations of Tat have been reported in the blood of HIV-1 infected people.<ref>Xiao ''et al. Proc. Natl. Acad. Sci. USA.'' '''2000'''. 97, 11466-11471.</ref> When the protein enters non-infected cells, the transcription efficiency increases. It is estimated that transcription levels are 10 to 100 fold higher during the transcription elongation stage when compared to normal basal transcription.<ref>Ensoli ''et al. Nature.'' '''1990'''. 345, 84-86.</ref> Green and Lowenstein, and Frankel and Pabo independently published studies in 1988 that demonstrated that Tat had the ability to cross cellular membranes and initiate transcription of HIV dsRNA.<ref>Green ''et al. Cell.'' '''1988'''. 55, 1179-1188.</ref><ref>Frankel ''et al. Cell.'' '''1988'''. 55, 1189-1193.</ref> These were the first known reports of a '''cell penetrating peptide (CPP)'''. |
| - | + | ||
| - | + | == '''Cell Penetrating Peptides''' == | |
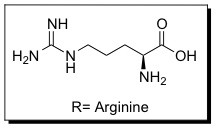
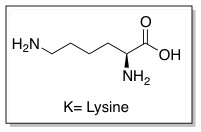
| - | + | '''Cell penetrating peptides (CPPs)''' are proteins with the ability to cross cellular membranes and facilitate the uptake of various cargo, such as small molecules, siRNA, and small DNA fragments.<ref>Futaki ''et al. J. Biol. Chem.'' '''2001'''. 276, 5836-5840.</ref> Such cargo can be associated via covalent or non-covalent interactions. Tat is considered a CPP because it contains a protein transduction domain (PTD). PTDs are cation-rich sequences of 10-30 residues, usually containing several Lysine and/or Arginine residues.<ref>Wender ''et al. Adv. Drug Deliv. Rev.'' '''2008'''. 60, 452-472.</ref> | |
| - | + | [[Image:Arginine1.jpg]] [[Image:Lysine1.jpg]] | |
| - | == | + | These sequences improve interactions between CPPs and cellular membranes and can help them enter cells. The (amino acid <scene name='Brittany_deRonde/Sandbox_1/Hiv_tat/3'>residues 47-57</scene>) of HIV-1 Tat comprise the protein transduction region of this protein. This arginine rich, 11 amino-acid sequence, <scene name='47/475184/Tat_arg/3'>YGRKKRRQRRR</scene> is now referred to as the TAT peptide, and has be shown to have improved cellular uptake compared to Tat. |
| - | ===Primary & Secondary Structure=== | ||
| - | {{STRUCTURE_1ema | SIZE=400 |PDB=1ema | SCENE=Green_Fluorescent_Protein/1ema_gfp_default/2 |CAPTION = 1ema, resolution 1.90Å (<scene name='Green_Fluorescent_Protein/1ema_gfp_default/2'>default scene</scene>).}} | ||
| - | Green fluorescent protein (<scene name='Green_Fluorescent_Protein/1ema_gfp_default/2'>default scene</scene>) is a 21 kDa protein consisting of 238 residues strung together<ref>Primary structure at [http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=1ema&template=protein.html&r=wiring&l=1&chain=A www.ebi.aci.uk].</ref> to form a | ||
| - | <scene name='Green_Fluorescent_Protein/1ema_gfp_barrel/2'>secondary structure</scene> of five α-helices and one eleven-stranded β-pleated sheet,<ref name="PDBsum" /> where each strand contains nine to thirteen residues each.<ref name="Ormo" /> (To view the primary and secondary structure of GFP, go to .) These β-strands display an almost “seamless symmetry” in which only two of the strands vary in structural content.<ref name="Phillips">PMID: 9434902</ref> This β-sheet conforms itself through regular hydrogen bonding into a β-barrel.<ref name="Yang" /> In GFP, the structure is so regular that <scene name='Green_Fluorescent_Protein/Water_stripes/1'>"stripes"</scene> of water molecules (red) can be seen following the structure of the barrel.<ref name="Phillips" /> Together with the α-helices at either end of the molecule, a nearly perfect cylinder is produced, 42Å long and 24Å in diameter,<ref name="Ormo" /> creating what is referred to as a “β-can” formation.<ref name="Phillips" /> The short helical segments at either end of the cylinder form “caps” to further protect the interior of the β-barrel.<ref name="Phillips" /> Overall stability is maintained by this β-can structure, helping to resist unfolding from heat and other denaturants.<ref name="Yang" /> | ||
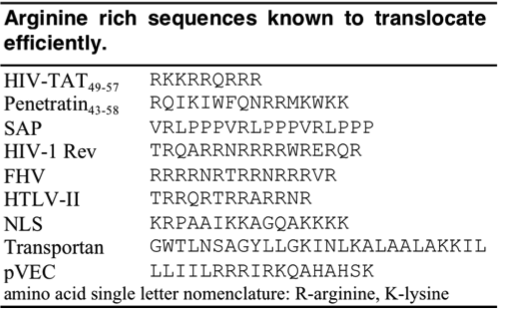
| - | + | Since the discovery of HIV Tat, other Arginine-rich, natural CPPs have been discovered including, penetratin from Drosophila antennapedia protein, sweet arrow peptide (SAP), HIV-1 Rev, flock house virus (FHV) coat, brome mosaic virus (BMV) Gag, human T-cell lymphotrophic virus (HTLV)-II Rex, and the nuclear localization signal (NLS) from nucleoplasmin. <ref>Futaki ''et al. J. Biol. Chem.'' '''2001'''. 276, 5836-5840.</ref><ref>Prochiantz ''et al. Adv. Drug Deliv. Rev.'' '''2008'''. 60, 448-451.</ref> The PTD sequences for these proteins can be found in the table below. | |
| - | {{Link Toggle FancyCartoonHighQualityView}}. | ||
| - | + | [[Image:Arginine_Rich_Peptides.png]] | |
| - | + | == '''Tat-mediated Transduction''' == | |
| - | + | The PTD in HIV Tat is highly cationic. When charged residues (Arginine or Lysine) are replaced with neutral residues (Alanine), cellular uptake decreases. It is likely that the cationic charge is necessary for electrostatic interactions with components of the cellular membrane, such as lipid head groups and proteoglycans.<ref>Ziegler ''et al. Biochemistry'' '''2003'''. 42, 9185-9194.</ref><ref>Goncalves ''et al. Biochemistry.'' '''2005'''. 44, 2692-2702.</ref> Other interactions may also play a role in interactions with the cellular membrane, such as hydrophobic and non-covalent interactions.<ref>Ziegler ''et al. Adv. Drug Deliv. Rev.'' '''2008'''. 60, 580-597.</ref> Although cellular uptake is not directly dependent upon architecture (branched vs. linear), it is dependent upon the number of Arginine residues. Polymers with less than five arginine residues are unable to enter cells.<ref>Wender ''et al. Proc. Natl. Acad. Sci.'' '''2000'''. 97, 13003-13008.</ref> As the number of residues increases, cellular uptake improves up until 15 residues where polymers can enter cells but with hindered efficiencies.<ref>Mitchell ''et al. J. Pept. Res.'' '''2000'''. 56, 318-325.</ref><ref>Futaki ''et al. J. Biol. Chem.'' '''2001'''. 276, 5836-5840.</ref> The exact mechanism of cellular uptake is still highly debated in the literature, but a couple of the proposed methods are discussed below. | |
| - | + | ||
| - | + | ==Transduction== | |
| + | Early studies indicated that HIV Tat primarily entered cells through transduction, meaning that the protein directly crossed the cell membranes. However, for most of these studies, cells were fixed with formaldehyde prior to sample analysis. In 2003, it was discovered that by fixing cells prior to analysis, it permeablized the cell membranes and resulted in artificially high cellular uptake values. Fixed cells showed higher concentrations of Tat in nuclei compared to non-fixed cells where it was localized in the cytoplasm. <ref>Richard ''et al. J. Biol. Chem.'' '''2003'''. 278, 585-590.</ref> Studies also showed that FACS could not tell the difference between internalized cells and those proteins that were only bound to the cellular membranes. Cellular uptake studies performed at 4°C and under conditions of ATP depletion show that a small percentage of CPPs can enter cells and also suggests that a majority of the protein enters cells through energy dependent pathways.<ref>Drin ''et al. J. Biol. Chem.'' '''2003'''. 276, 31192-31201.</ref> Some groups have also studied transduction of Tat using model vesicles. <ref>Ziegler ''et al. Biochemistry.'' '''2003'''. 278, 9185-9194.</ref> | ||
| - | + | ==Endocytosis== | |
| + | Another proposed mechanism for cellular uptake is endocytosis, specifically macropinocytosis. This form of fluid phase endocytosis is less known that other types (phagocytosis, clathrin, and coated pit), it is believed to occur in all cell types and enable the uptake of large molecules. <ref>Gump ''et al. Trends in Molecular Medicine.'' '''2007'''. 13, 443-448.</ref> During this process, actin protrusions fold around molecules and the surrounding medium and enable cellular uptake. | ||
| - | + | == '''My Research Interests''' == | |
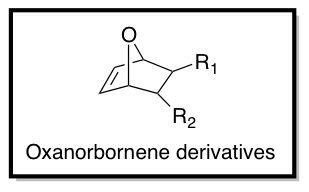
| + | As part of the [http://www.pse.umass.edu/gtew/index.html Tew Research Group] in the [http://www.pse.umass.edu Department of Polymer Science and Engineering] at the University of Massachusetts, Amherst, I am interested in the design of Tat-inspired CPPs for drug and gene delivery. Using functionalized oxanorbornene derivatives as monomers (as shown below), ring opening metathesis polymerization (ROMP) is performed using Grubbs' 3rd generation catalyst to synthesize cationic CPP mimics. Hydrophobicity, charge, aromaticity, and pi-electronics of the monomer structures can be optimized to achieve better cellular uptake and cell viability by changing R1 and R2. | ||
| - | + | [[Image:norbornenederivatives.png]] | |
| - | + | ||
| - | + | == '''Acknowledgments''' == | |
| - | + | The Tew research group is gratefully acknowledged for their contributions to the design and content of this page. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | == '''References''' == | ||
{{Reflist}} | {{Reflist}} | ||
| - | |||
| - | ==Additional Resources== | ||
| - | *For additional information, see: [[Colored & Bioluminescent Proteins]] | ||
| - | *[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1ema First Glance] | ||
| - | *PDBsum: [http://www.ebi.ac.uk/pdbsum/1ema 1ema] | ||
| - | *RCSB PDB [http://www.rcsb.org/pdb/explore.do?structureId=1ema 1ema] | ||
| - | *[http://oca.weizmann.ac.il/oca-bin/ocaids?id=1ema OCA] | ||
| - | *UniProt: [http://www.uniprot.org/uniprot/P42212 P42212] | ||
| - | *Scop: [http://scop.mrc-lmb.cam.ac.uk/scop/data/scop.b.e.gc.b.b.b.html P42212] | ||
| - | *CATH: [http://www.cathdb.info/domain/1emaA00 1emaA00] | ||
| - | *Pfam: [http://pfam.sanger.ac.uk/family?acc=PF01353 PF01353] | ||
| - | *InterPro: [http://www.ebi.ac.uk/interpro/ISearch?query=IPR000786 IPR000786] | ||
| - | *[http://www.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/pdb42_1.html GFP featured] at the '''[http://www.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month]''' series of tutorials by [[User:David_S._Goodsell|David Goodsell]]. | ||
| - | * [http://www.nature.com/nature/journal/v462/n7270/edsumm/e091112-05.html Inside green fluorescent protein] - editor's summary that accompanied [http://www.nature.com/nature/journal/v462/n7270/covers/ structural detail of GFP chromophore on the cover] of Nature. | ||
| - | |||
| - | [[he:GFP_(Hebrew)]] | ||
| - | |||
| - | [[Category:Topic Page]] | ||
Current revision
One of the CBI Molecules being studied in the University of Massachusetts Amherst Chemistry-Biology Interface Program at UMass Amherst and on display at the Molecular Playground.
Contents |
Introduction
HIV Tat, or simply Tat, is a human immunodeficiency virus (HIV) gene that regulates transcription of HIV dsRNA.[1] Tat, which stands for trans-activator of transcription, contains 86 amino acid residues in its sequence.[2][3]
|
Cell Penetrating Peptides
Cell penetrating peptides (CPPs) are proteins with the ability to cross cellular membranes and facilitate the uptake of various cargo, such as small molecules, siRNA, and small DNA fragments.[10] Such cargo can be associated via covalent or non-covalent interactions. Tat is considered a CPP because it contains a protein transduction domain (PTD). PTDs are cation-rich sequences of 10-30 residues, usually containing several Lysine and/or Arginine residues.[11]
These sequences improve interactions between CPPs and cellular membranes and can help them enter cells. The (amino acid ) of HIV-1 Tat comprise the protein transduction region of this protein. This arginine rich, 11 amino-acid sequence, is now referred to as the TAT peptide, and has be shown to have improved cellular uptake compared to Tat.
Since the discovery of HIV Tat, other Arginine-rich, natural CPPs have been discovered including, penetratin from Drosophila antennapedia protein, sweet arrow peptide (SAP), HIV-1 Rev, flock house virus (FHV) coat, brome mosaic virus (BMV) Gag, human T-cell lymphotrophic virus (HTLV)-II Rex, and the nuclear localization signal (NLS) from nucleoplasmin. [12][13] The PTD sequences for these proteins can be found in the table below.
Tat-mediated Transduction
The PTD in HIV Tat is highly cationic. When charged residues (Arginine or Lysine) are replaced with neutral residues (Alanine), cellular uptake decreases. It is likely that the cationic charge is necessary for electrostatic interactions with components of the cellular membrane, such as lipid head groups and proteoglycans.[14][15] Other interactions may also play a role in interactions with the cellular membrane, such as hydrophobic and non-covalent interactions.[16] Although cellular uptake is not directly dependent upon architecture (branched vs. linear), it is dependent upon the number of Arginine residues. Polymers with less than five arginine residues are unable to enter cells.[17] As the number of residues increases, cellular uptake improves up until 15 residues where polymers can enter cells but with hindered efficiencies.[18][19] The exact mechanism of cellular uptake is still highly debated in the literature, but a couple of the proposed methods are discussed below.
Transduction
Early studies indicated that HIV Tat primarily entered cells through transduction, meaning that the protein directly crossed the cell membranes. However, for most of these studies, cells were fixed with formaldehyde prior to sample analysis. In 2003, it was discovered that by fixing cells prior to analysis, it permeablized the cell membranes and resulted in artificially high cellular uptake values. Fixed cells showed higher concentrations of Tat in nuclei compared to non-fixed cells where it was localized in the cytoplasm. [20] Studies also showed that FACS could not tell the difference between internalized cells and those proteins that were only bound to the cellular membranes. Cellular uptake studies performed at 4°C and under conditions of ATP depletion show that a small percentage of CPPs can enter cells and also suggests that a majority of the protein enters cells through energy dependent pathways.[21] Some groups have also studied transduction of Tat using model vesicles. [22]
Endocytosis
Another proposed mechanism for cellular uptake is endocytosis, specifically macropinocytosis. This form of fluid phase endocytosis is less known that other types (phagocytosis, clathrin, and coated pit), it is believed to occur in all cell types and enable the uptake of large molecules. [23] During this process, actin protrusions fold around molecules and the surrounding medium and enable cellular uptake.
My Research Interests
As part of the Tew Research Group in the Department of Polymer Science and Engineering at the University of Massachusetts, Amherst, I am interested in the design of Tat-inspired CPPs for drug and gene delivery. Using functionalized oxanorbornene derivatives as monomers (as shown below), ring opening metathesis polymerization (ROMP) is performed using Grubbs' 3rd generation catalyst to synthesize cationic CPP mimics. Hydrophobicity, charge, aromaticity, and pi-electronics of the monomer structures can be optimized to achieve better cellular uptake and cell viability by changing R1 and R2.
Acknowledgments
The Tew research group is gratefully acknowledged for their contributions to the design and content of this page.
References
- ↑ Sodroski et al. Science. 1985. 227, 171-173. [1] http://www.jstor.org/stable/1695050
- ↑ Arya et al. Science 1985. 229, 69-73.
- ↑ Sodroski et al. Science. 1985. 229, 74-77.
- ↑ Dayton et al. Cell. 1986. 44, 941-947.
- ↑ Fisher et al. Nature. 1986. 320, 367-371.
- ↑ Xiao et al. Proc. Natl. Acad. Sci. USA. 2000. 97, 11466-11471.
- ↑ Ensoli et al. Nature. 1990. 345, 84-86.
- ↑ Green et al. Cell. 1988. 55, 1179-1188.
- ↑ Frankel et al. Cell. 1988. 55, 1189-1193.
- ↑ Futaki et al. J. Biol. Chem. 2001. 276, 5836-5840.
- ↑ Wender et al. Adv. Drug Deliv. Rev. 2008. 60, 452-472.
- ↑ Futaki et al. J. Biol. Chem. 2001. 276, 5836-5840.
- ↑ Prochiantz et al. Adv. Drug Deliv. Rev. 2008. 60, 448-451.
- ↑ Ziegler et al. Biochemistry 2003. 42, 9185-9194.
- ↑ Goncalves et al. Biochemistry. 2005. 44, 2692-2702.
- ↑ Ziegler et al. Adv. Drug Deliv. Rev. 2008. 60, 580-597.
- ↑ Wender et al. Proc. Natl. Acad. Sci. 2000. 97, 13003-13008.
- ↑ Mitchell et al. J. Pept. Res. 2000. 56, 318-325.
- ↑ Futaki et al. J. Biol. Chem. 2001. 276, 5836-5840.
- ↑ Richard et al. J. Biol. Chem. 2003. 278, 585-590.
- ↑ Drin et al. J. Biol. Chem. 2003. 276, 31192-31201.
- ↑ Ziegler et al. Biochemistry. 2003. 278, 9185-9194.
- ↑ Gump et al. Trends in Molecular Medicine. 2007. 13, 443-448.