What is a Riboswitch?
Normally, a variety of proteins and protein cofactors control gene expression in an organism by binding to different sites on messenger RNA (mRNA). Riboswitches are genetic regulatory elements that are built directly into the RNA. They are a type of noncoding RNA that regulate gene expression in the absence of proteins by switching from one structural conformation (shape) to another in response to ligand binding[1]. Most contain a single binding site that recognizes a specific ligand. The ability of a riboswitch to discriminate against molecules that are similar or closely related to its ligand is essential to prevent metabolic misregulation[2].
The various classes of riboswitches discovered so far are differentiated by their respective ligands. Every class of riboswitch is characterized by an aptamer (binding site) domain, which provides the site for ligand binding, and an expression platform that undergoes conformational change. The sequences and structures of aptamer domains are highly conserved, and therefore exhibit little variation among riboswitches belonging to the same class[1].
Atomic-resolution structures of riboswitch binding sites show that they make numerous hydrogen bonds with their ligands, forming contacts that stabilize RNA interactions to further increase affinity. Some binding sites form pockets that entirely engulf the ligand, and in these instances an induced-fit mechanism of binding must occur[2].
Molecular Structure
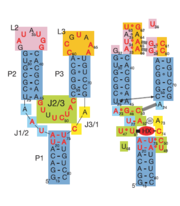
Fig. 1: Left: Secondary structure of riboswitch. Right: Riboswitch with bound ligand
The structure of this riboswitch consists of three , and , that surround a . This junction defines the binding pocket, which is formed by two sets of base triples that flank the site from above and below[3].
In the experiment described by the abstract above, hypoxanthine binds to the junction via a series of hydrogen bonds with nucleotides . The mRNA contacts all of the functional groups in hypoxanthine, which explains the specificity of the riboswitch for hypoxanthine. Additionally, the two carbonyl oxygens located at the 2-position of are able to form hydrogen bonds with guanine's exocyclic amino group
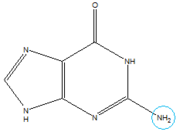
Structure of Guanine with exocyclic amino group circled in blue.
. This gives the riboswitch a tenfold higher affinity for guanine over hypoxanthine
[3].
When the riboswitch is in a ligand-bound state, the RNA to forms a three-dimentional fold in which the terminal loops become connected through a series of hydrogen bonds (Fig. 1). This causes the and helices to become parallel to each other. The unfavorable electrostatic interactions of the parallel ribose-phosphate backbones are neutralized through the binding multiple Co(NH
3)
63+ between the backbones. The interactions between the terminal loops stabilize the tertiary structure of the riboswitch, and are essential for ligand binding to occur. Experiments in which the loops are replaced with extremely stable UUCG sequences (Fig. 2) have demonstrated that alterations to the terminal loops prevent the loop-loop interactions from forming, and eliminate the riboswitch's ability to recognize hypoxanthine as a result.
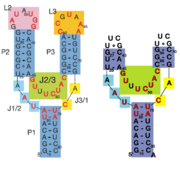
Fig. 2: Left: Riboswitch with wild-type terminal loops. Right: Riboswitch with terminal loops replaced by UUCG tetraloops.
As such, this tertiary interaction is required for ligand recognition, even though it never contacts the ligand directly
[3].
When bound, the ligand is almost completely enveloped by the RNA (Fig. 3).
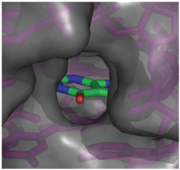
Fig. 3: Molecular surface representation of the binding pocket of the guanine riboswitch bound to hypoxanthine.
This implies that the binding site must undergo significant conformational change upon ligand binding, as a preformed binding site would inhibit the ligand's access to it
[3].
Gene Regulation
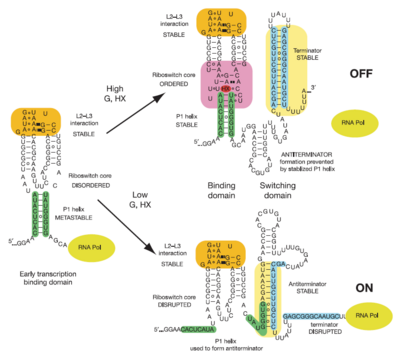
This riboswitch acts as a transcriptional regulation mechanism in the operon of the xpt-pbuX purine salvaging gene of B. subtilis, and directly represses gene expression in response to an increase in intracellular concentrations of guanine, xanthine, or hypoxanthine. Transcription of the initial ~90 nucleotides of the sequence triggers the interaction of the loops. This results in the organization of the such that it is partially unstructured to allow access to the site while still allowing efficient ligand binding.
Illustrated below are the conformational changes made by the riboswitch in response to changes in ligand concentration. At low concentrations of ligand (in this case hypoxanthine), the becomes destabilized. This allows the RNA to incorporate part of the helix into an antiterminator element as illustrated below, resulting in continued transcription. At sufficiently high concentrations of ligand, the nucleobase binds to the junction and stabilizes the . This prevents the helix from being incorporated into an antiterminator element, resulting in the formation of a stable stem-loop structure that halts transcription via Rho-independent termination[3].
Potential Applications of Riboswitches
Riboswitches in general have various important applications. They can be useful in gaining valuable insights into how gene regulation mechanisms have evolved from the primitive forms of life to the more complex ones. Riboswitches have also been used as potential drug targets for antibacterial and antifungal agents, as they play important roles in controlling the exrpession of numerous genes involved in metabolism and transport processes[1].
Artificial riboswitches have also been engineered for the manipulation of gene expression. Identifying the principles of riboswitch-mediated regulation may lead to the development of engineered ligands capable of modulating gene expression. More detailed characterization of the distribution and function of riboswitches across and within different genomes is essential to determine their precise role as riboregulators and potential drug targets[1].
3D structures of riboswitch
Riboswitch






