Yersinia YopH
From Proteopedia
(Difference between revisions)
| (8 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
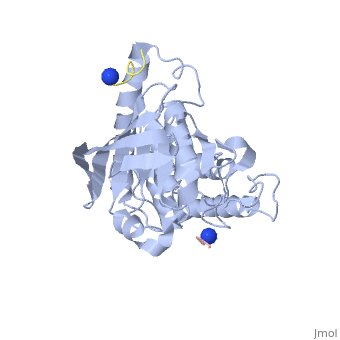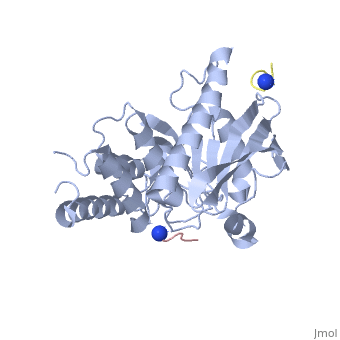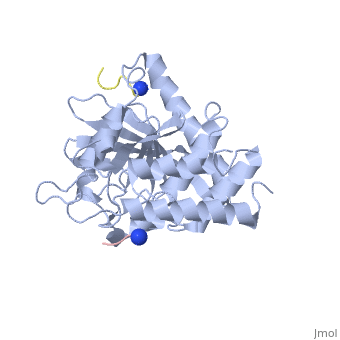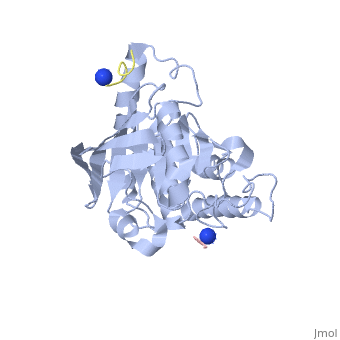
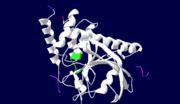
| - | < | + | <StructureSection load='1xxv' size='350' side='right' scene='' caption='YopH (grey, green) complex with EGFR peptide (pink, yellow, magenta, cyan) and NH2 groups (PDB code [[1xxv]])'> |
| - | [[Yersinia YopH]] (Yop51) is a virulence factor of bacteria of the genus ''Yersinia'', which includes ''Y. pestis'' (which can manifest in one form as bubonic plague), ''Y. pseudotuberculosis'', and ''Y. enterocolitica''. YopH is a protein tyrosine phosphatase (PTPase) which is injected into infected cells by a type III secretion mechanism<ref name="Ivanov">Ivanov, M.I., Stuckey, J.A., Schubert, H.L. Saper, M.A., Bliska, J.B. Two substrate-targeting sites in the ''Yersinia'' protein tyrosine phosphatase co-operate to promote bacterial virulence. ''Molecular Microbiology''. 2005. 55:1346-1356.</ref><ref name="Persson"/><ref name="Black"> Black, D.S., and Bliska, J.B. Identification of p130Cas as a substrate of ''Yersinia'' YopH(Yop51), a bacterial protein tyrosine phosphatase that translocates into mammalian cells and targets focal adhesions. ''The EMBO Journal''. 1997. 16:2730-2744.</ref> | + | |
| + | [[Yersinia YopH]] (Yop51) is a virulence factor of bacteria of the genus ''Yersinia'', which includes ''Y. pestis'' (which can manifest in one form as bubonic plague), ''Y. pseudotuberculosis'', and ''Y. enterocolitica''. YopH is a protein tyrosine phosphatase (PTPase) which is injected into infected cells by a type III secretion mechanism<ref name="Ivanov">Ivanov, M.I., Stuckey, J.A., Schubert, H.L. Saper, M.A., Bliska, J.B. Two substrate-targeting sites in the ''Yersinia'' protein tyrosine phosphatase co-operate to promote bacterial virulence. ''Molecular Microbiology''. 2005. 55:1346-1356.</ref><ref name="Persson"/><ref name="Black"> Black, D.S., and Bliska, J.B. Identification of p130Cas as a substrate of ''Yersinia'' YopH(Yop51), a bacterial protein tyrosine phosphatase that translocates into mammalian cells and targets focal adhesions. ''The EMBO Journal''. 1997. 16:2730-2744.</ref> <scene name='39/399846/Cv/1'>Crystal structure showing two YopH protein-tyrosine phosphatase catalytic domains from Yersinia enterocolitica bound to EGFR-derived peptides</scene> ([[1xxv]]). | ||
== Yersina Outer Proteins == | == Yersina Outer Proteins == | ||
| Line 12: | Line 13: | ||
== YopH Structure == | == YopH Structure == | ||
| - | YopH is a 51kDa, 468 amino acid protein with two domains linked by a proline rich sequence<ref name="Ivanov" /><ref name="Cornelis"/>. Residues 1-129 form the N-terminal domain which contains a binding site ("site 1") for substrate targeting<ref name="Ivanov" />. This site is not catalytic, and the binding domain appears to be unique among phosphotyrosine binding domains as it does not resemble SH2 (Src homology 2) or PTB (phospotyrosine-binding) domains<ref name="Ivanov" />. The second domain is the catalytically active domain with a mixed β-sheet in the center of the protein, surrounded by α-helices<ref name="Stuckey" />. This domain encompasses the residues from approximately 163-468, and has two binding sites<ref name="Ivanov" />. One appears to be involved in substrate specificity ("site 2"), while the other is the catalytic site<ref name="Ivanov" />. | + | YopH is a 51kDa, 468 amino acid protein with two domains linked by a proline rich sequence<ref name="Ivanov" /><ref name="Cornelis"/>. Residues 1-129 form the N-terminal domain which contains a binding site ("site 1") for substrate targeting<ref name="Ivanov" />. This site is not catalytic, and the binding domain appears to be unique among phosphotyrosine binding domains as it does not resemble SH2 (Src homology 2) or PTB (phospotyrosine-binding) domains<ref name="Ivanov" />. The second domain is the catalytically active domain with a mixed β-sheet in the center of the protein, surrounded by α-helices<ref name="Stuckey" />. This domain encompasses the residues from approximately 163-468, and has two binding sites<ref name="Ivanov" />. One appears to be involved in substrate specificity ("site 2"), while the other is the catalytic site<ref name="Ivanov" />. |
| - | + | [[Image:Catalytic domain of YopH.gif|thumb|left|The catalytic domain of YopH. The C-terminal residue is coloured in red, the residue closest to the N-terminal of the protein is coloured in blue, the phosphate binding loop is coloured green, and the phosphopeptide ligands are purple. ]] | |
| - | + | {{Clear}} | |
| - | [[ | + | <scene name='Sandbox_162/Nterminal_subunit_yoph/2'> The N-terminal subunit of YopH</scene> ([[1k46]]) is important for secretion, binding of SycH, and binding of the substrates to the catalytic domain. This domain contains four α-helices and four β-sheets in an α+β arrangement<ref name="Smith"/>. The amino-terminal α-helix (residues 2-17) makes up the secretion signal<ref name="Smith">Smith, C.L., Khandelwal, P., Keliikuli, K., Zuiderweg, E.R.P. and Saper, M. Structure of the type III secretion and substrate-binding domain of ''Yersinia'' YopH phosphatase. ''Molecular Microbiology''. 2001. 42:967-979.</ref>. Since there does not appear to be a conserved signal sequence between secreted proteins in type III secretion systems, nor a signal peptide cleaved, it has been suggested that recognition of secreted proteins may be by a structural motif, such as an amphipathic helix<ref name="Smith" />. The chaperone SycH binds to residues 20-70 in a hydrophobic patch from a nascent or partially unfolded N-terminal domain<ref name="Smith" />. <br/> |
| + | |||
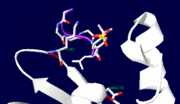
| + | The <scene name='Sandbox_162/Catalytic_subunit_yopha/1'>catalytic domain</scene> (1xxv), contains seven α-helices and a mixed β-sheet with eight strands, with two βαβ motifs in the center<ref name="Stuckey"/>. X-ray crystallography (using a phosphopeptide substrate which resembles the autophosphorylation site on EGFR) suggests that there are two substrate binding sites on this domain. Site 2 appears be more specific than the active site as it binds 5-6 residues, while the active site only binds 3-4 of these substrate residues <ref name="Ivanov" />. In site 2, the phosphotyrosine binds with its ring parallel to the protein surface, and it forms salt bridges with two residues, Arg 278 (salt bridge to the phosphate) and Lys 342 (salt bridge to: phosphonate oxygens, fluorine atom, and Glu4 of substrate)<ref name="Ivanov" />. | ||
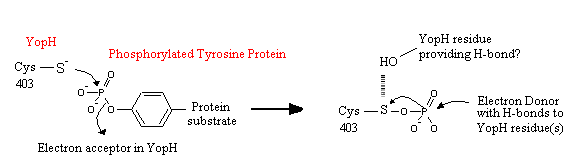
In the active site, residues 403-410 form a phosphate binding loop (P-loop)<ref name="Ivanov" />. The first residue in this loop is crucial for enzyme activity. If the cysteine at position 403 (<scene name='Sandbox_162/Catalytic_subunit_yophacts/1'>Cys403</scene>) is substituted with serine,the enzyme will be catalytically inactive and will trap the substrate<ref name="Ivanov" />. Another crucial residue also lies within this P-loop. If arginine 409 is substituted to serine, substrate binding and catalysis is inhibited (it does not trap the substrate like the Cys 403 mutation to Serine)<ref name="Ivanov" />. | In the active site, residues 403-410 form a phosphate binding loop (P-loop)<ref name="Ivanov" />. The first residue in this loop is crucial for enzyme activity. If the cysteine at position 403 (<scene name='Sandbox_162/Catalytic_subunit_yophacts/1'>Cys403</scene>) is substituted with serine,the enzyme will be catalytically inactive and will trap the substrate<ref name="Ivanov" />. Another crucial residue also lies within this P-loop. If arginine 409 is substituted to serine, substrate binding and catalysis is inhibited (it does not trap the substrate like the Cys 403 mutation to Serine)<ref name="Ivanov" />. | ||
| Line 23: | Line 26: | ||
== Function == | == Function == | ||
| - | + | [[Image:Key_residues_at_site_2-Arg_278_and_Lys_342.gif|thumb|left|Key residues involved in forming salt-bridges with the substrate in site 2 on the catalytic subunit]] During infection, the Yersinia bacteria are attached to the host cells, but cannot be ingested. In order to prevent this, protein tyrosine kinase activity in the host cell must be inhibited<ref name="Persson"/>. Since focal adhesion (locations where the actin cytoskeleton is linked to the extracellular matrix) assembly is part of the signal cascade involved in the process of clustering of β-1 integrins and tyrosine kinase activity in response to pathogens, the protein tyrosine phosphatase (PTPase) activity of YopH could disrupt this interaction and thus prevent uptake of the bacteria<ref name="Persson"/>. Focal adhesion kinase (FAK) and p130 Cas are both phosphorylated in response to the integrin-mediated focal adhesion assembly. YopH has been shown to dephosphorylate both FAK and p130 Cas with a high specificity, thereby blocking the uptake of the bacteria by the host cell ("anti-phagocytosis")<ref name="Persson"/><ref name="Black"/>. YopH has also been shown to interact with and dephosphorylate Fyn-binding protein<ref name="Hamid"/>. | |
| - | During infection, the Yersinia bacteria are attached to the host cells, but cannot be ingested. In order to prevent this, protein tyrosine kinase activity in the host cell must be inhibited<ref name="Persson"/>. Since focal adhesion (locations where the actin cytoskeleton is linked to the extracellular matrix) assembly is part of the signal cascade involved in the process of clustering of β-1 integrins and tyrosine kinase activity in response to pathogens, the protein tyrosine phosphatase (PTPase) activity of YopH could disrupt this interaction and thus prevent uptake of the bacteria<ref name="Persson"/>. Focal adhesion kinase (FAK) and p130 Cas are both phosphorylated in response to the integrin-mediated focal adhesion assembly. YopH has been shown to dephosphorylate both FAK and p130 Cas with a high specificity, thereby blocking the uptake of the bacteria by the host cell ("anti-phagocytosis")<ref name="Persson"/><ref name="Black"/>. YopH has also been shown to interact with and dephosphorylate Fyn-binding protein<ref name="Hamid"/>. | + | |
== Mechanism == | == Mechanism == | ||
| Line 35: | Line 37: | ||
The mechanism proposed here is only the most commonly suggested mechanism. Several studies were unable to prove a direct interaction between YopH and Cas or Fyn-binding protein, nor were they able to confirm the general mechanism by which YopH inhibits phagocytosis of the bacterium<ref name="Hamid"/><ref name="Vinney">DeVinney, R., Steele-Mortimer, O., and Finlay, B.B. Phosphatases and kinases delivered to the host cell by bacterial pathogens. ''Trends in Microbiology''. 2000. 8:29-33.</ref>. For instance, it has not been confirmed that several of YopH's substrates are tyrosine-phosphorylated as part of the integrin-mediated response to binding of the bacterium, and thus it may have a different mechanism of action<ref name="Hamid"/><ref name="Vinney"/>. | The mechanism proposed here is only the most commonly suggested mechanism. Several studies were unable to prove a direct interaction between YopH and Cas or Fyn-binding protein, nor were they able to confirm the general mechanism by which YopH inhibits phagocytosis of the bacterium<ref name="Hamid"/><ref name="Vinney">DeVinney, R., Steele-Mortimer, O., and Finlay, B.B. Phosphatases and kinases delivered to the host cell by bacterial pathogens. ''Trends in Microbiology''. 2000. 8:29-33.</ref>. For instance, it has not been confirmed that several of YopH's substrates are tyrosine-phosphorylated as part of the integrin-mediated response to binding of the bacterium, and thus it may have a different mechanism of action<ref name="Hamid"/><ref name="Vinney"/>. | ||
| + | </StructureSection> | ||
| + | ==3D structures of protein tyrosine phosphatase== | ||
| + | |||
| + | [[Protein tyrosine phosphatase]] | ||
==References== | ==References== | ||
<references /> | <references /> | ||
Current revision
| |||||||||||
3D structures of protein tyrosine phosphatase
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 Ivanov, M.I., Stuckey, J.A., Schubert, H.L. Saper, M.A., Bliska, J.B. Two substrate-targeting sites in the Yersinia protein tyrosine phosphatase co-operate to promote bacterial virulence. Molecular Microbiology. 2005. 55:1346-1356.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Persson, C. Carballeira, N., Wolf-Watz, H., and Fällman, M. The PTPase YopH inhibits uptake of Yersinia, tyrosine phosphorylation of pl30cas and FAK, and the associated accumulation of these proteins in peripheral focal adhesions. The EMBO Journal. 1997. 16:2307-2318.
- ↑ 3.0 3.1 Black, D.S., and Bliska, J.B. Identification of p130Cas as a substrate of Yersinia YopH(Yop51), a bacterial protein tyrosine phosphatase that translocates into mammalian cells and targets focal adhesions. The EMBO Journal. 1997. 16:2730-2744.
- ↑ 4.0 4.1 4.2 4.3 4.4 Hamid, N., Gustavsson, A.,Andersson, K., McGee, K., Persson, C., Rudd, C.E., Fällman, M. YopH dephosphorylates Cas and Fyn-binding protein in macrophages. Microbial Pathogenesis. 1999. 26:231-242
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 Galán, J.E. and Collmer, A. Type III Secretion Machines: Bacterial Devices for Protein Delivery into Host Cells. Science. 1999. 284:1322-1328
- ↑ 6.0 6.1 Wulff-Strobel, C.R., Williams, A.W., Straley, S.C. LcrQ and SycH function together at the Ysc type III secretion system in Yersinia pestis to impose a hierarchy of secretion. Molecular Microbiology.2002. 43:411-423
- ↑ 7.0 7.1 7.2 Cornelis, G.R. and Wolf-Watz, H. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Molecular Microbiology. 1997. 23:861-867.
- ↑ 8.0 8.1 8.2 8.3 . Stuckey, J.A., Schubert, H.L., Fauman, E.B., Zhang, Z., Dixon, J.E., and Saper, M.A. Crystal structure of Yersinia protein tyrosine phosphatase at 2.5Å and the complex with tungstate. Nature. 1994. 370:571-575.
- ↑ 9.0 9.1 9.2 9.3 Smith, C.L., Khandelwal, P., Keliikuli, K., Zuiderweg, E.R.P. and Saper, M. Structure of the type III secretion and substrate-binding domain of Yersinia YopH phosphatase. Molecular Microbiology. 2001. 42:967-979.
- ↑ 10.0 10.1 Pannifer, A.D.B., Flint, F.J., Tonks, N.K., and Barford, D. Visualization of the Cysteinyl-phosphate Intermediate of a Protein-tyrosine Phosphatase by X-ray Crystallography. The Journal of Biological Chemistry. 1998. 273:10454-10462.
- ↑ 11.0 11.1 DeVinney, R., Steele-Mortimer, O., and Finlay, B.B. Phosphatases and kinases delivered to the host cell by bacterial pathogens. Trends in Microbiology. 2000. 8:29-33.
Proteopedia Page Contributors and Editors (what is this?)
David Canner, Michal Harel, Cara Halseth, Alexander Berchansky