|
|
| (316 intermediate revisions not shown.) |
| Line 1: |
Line 1: |
| - | <!-- DO NOT DELETE THE TEMPLATE LINE --> | + | <references/><references/><!-- DO NOT DELETE THE TEMPLATE LINE --> |
| | {{Template:Sandbox Reserved Lynmarie Thompson}} | | {{Template:Sandbox Reserved Lynmarie Thompson}} |
| | <!-- INSERT YOUR SCENES AND TEXT BELOW THIS LINE --> | | <!-- INSERT YOUR SCENES AND TEXT BELOW THIS LINE --> |
| | | | |
| - | =='''Cisplatin-DNA complex- 1a84'''== | + | =='''P2Y12 Receptor in Complex with AZD1283 (4ntj)<ref>PMID: 24670650 </ref>'''== |
| | + | by [Cora Ricker, Lauren Timmins, Aidan Finnerty, Adam Murphy, Duy Nguyen] |
| | | | |
| - | ===Introduction===
| + | [[Student Projects for UMass Chemistry 423 Spring 2016]] |
| - | <Structure load='1a84' size='500' frame='true' align='right' caption='Cisplatin' scene='Insert optional scene name here' /> | + | <StructureSection load='4ntj' size='350' side='right' caption='Modeling of P2Y12 binded with Antithrombotic Drug([[4ntj]])' scene=''> |
| | | | |
| - | The figure to the right shows <scene name='Sandbox_Reserved_430/Cisplatin_intro_with_caption/2'>cisplatin</scene> bound to a 12 base pair double stranded DNA. Cisplatin, cis-PtCl2(NH3)2, is an “alkylating” chemotherapy drug, administered intravenously, used in the treatment of various types of cancer. <ref> adhttp://en.wikipedia.org/wiki/Chemotherapy</ref> | + | ==Introduction== |
| - |
| + | The goal of pharmaceuticals is to prevent or cure disease through drug therapy by specifically targeting cells, proteins, enzymes, genes, etc. It is often crucial to understand the structure, function, and relevant mechanisms involved with the target when designing an effective drug candidate. Furthermore, being able to know the effects on structure after drug-binding can provide insight into the functionality of a specific target. Because of this fact, it is common practice for research labs to develop protein crystals and use methods like x-ray diffraction, electron density mapping, and nuclear magnetic resonance spectroscopy to model the full structure. In this case, P2Y12, a member of P2Y receptor, was modeled when binded to AZD1283, an engineered receptor inhibitor.The molecular scene shows the chemical<scene name='48/483887/Here_is/1'> model</scene> of P2Y12 with the anionic side chains in red and charged nucleic acids and ligands in grey for contrast. |
| - | There are three fundamental components in the mechanism of cisplatin – cisplatin, DNA, and HMG-protein. Cisplatin makes contact with the cell membrane and enters the cell through active transport, but some molecules are passively diffused. This platinum-based drug acts in vivo by <scene name='Sandbox_Reserved_430/Guanine_in_black_caption/2'>binding</scene> to two consecutive adjacent '''guanine''' bases in DNA leading to the loss of its chlorine atoms for the nitrogen on the guanine; this occurs to better balance the platinum charge.<ref> David, G.S. The Molecular Perspective: Cisplatin. doi: 10.1634/theoncologist.11-3-316 The Oncologist March 2006 vol. 11 no. 3 316-317.</ref> The binding of cisplatin creates a 49<scene name='Sandbox_Reserved_430/49_bend_caption/1'>49°</scene> bend with an overall helix bend of 78<scene name='Sandbox_Reserved_430/78_bend/2'>78°</scene>, which is crucial to cisplatin’s role as an anticancer drug.[5] The bend in the <scene name='Sandbox_Reserved_430/Hmg-proetin_bound/1'>DNA</scene>, as seen in pdb 1ckt, allows for <font color='magenta'>HMG-protein</font> to bind to the DNA, and when bound it inserts a wedge like phenol group of '''phenylalanine''' <scene name='Sandbox_Reserved_430/37_phenylalanine/2'>37</scene> into the widened minor grove. HMG-proteins, high mobility group-proteins, are found everywhere and regulate transcription, replication, recombination and repair, and once bound to the DNA it de-stacks the <font color='cyan'>nucleotide base pairs </font>, which in turn kinks the already mutated DNA. With the HMG-protein bound to the DNA, the cell cannot properly repair the DNA, leading to apoptosis.<ref> Gelasco, Andrew. "NMR solution and structure of DNA Dodecamer Duplex Containing cis-Diammaineplatium" Department of Chemistry, MIT:1998</ref>
| + | |
| | | | |
| - | This link shows a video on the mechanism of cisplatin.[http://www.youtube.com/watch?v=Wq_up2uQRDo] | |
| | | | |
| - | Unfortunately, there is not yet a definitive way to regulate which cells are affected by cisplatin, so the cytotoxic effects damage normal cells as well, in particular rapidly dividing cells such as those found in the gastrointestinal tract, bone marrow, testicles, ovaries, and hair growth. It is the foundation to many combination treatments for cancers, but not all cancers are effected by cisplatin, the majority of patients using cisplatin will relapse with platinum resistant diseases. Another way cisplatin can be in effective, is when the cancers gets too old; when a tumor starts out it divides more frequently and this is when it is effected. This same logic goes for solid tumors, other treatments are needed for these types of issues.
| + | Image analysis of P2Y12 crystals is used to model protein structure in complex with AstraZeneca’s novel AZD1283: Ethyl 6-(4(-((benzylsulphonyl)carbamoyl)piperidin-1yl)-5-cyano-2-methylnicotinate. AZD1283 functions to block the P2Y12 receptor as a means to treat thrombosis. AZD1283-binding leads to unique protein structure, unfound in other P2Y receptors. Helix V of seven transmembrane helices is found to be <scene name='48/483887/Helix_vii/1'>elongated and straightened</scene>. This change along with the discovery of a potential second active within P2Y12 has implications on how P2Y12 uses it’s seven transmembrane helical bundle interact with ADP in the bloodstream. |
| | | | |
| | + | As a member of a large class of G-protein-coupled receptors, P2Y12 is often an initial role player in signal transduction and cellular response due to external environmental factors. In the case of P2Y12, the receptor responds to ADP concentrations in the extracellular matrix and on a larger scale the blood stream. There is importance is understanding how P2Y12's structure receives ADP as an activator. In turn, knowledge of how P2Y12 is affected when properly inhibited can lead to improved drug design in terms of bioavailability, binding affinity, and effectiveness of inhibition. Ultimately, pharmaceuticals will be better able to prevent and treat cardiovascular diseases and medical conditions (thrombosis, hypercoagulable states) and more immediate dangers (stroke, embolism, and heart attacks). |
| | + | ==Overall Structure== |
| | + | P2Y12R is a 1 chain structure. The <scene name='48/483887/Secondary_structure/2'>secondary structure</scene> of P2Y12R consists of eight <font color='deeppink'>alpha helices</font>. Seven transmembrane alpha helices are tilted and in a bundle, while the carboxy-terminal helix VIII is parallel to the membrane bilayer. The <scene name='48/483887/Rainbow/3'>rainbow scene</scene> demonstrates how the chain goes from the <font color='blue'>N</font> to <font color='red'>C</font> termini with each helix being approximately one color each of the color scheme. |
| | + | {{Template:ColorKey_N2CRainbow}} |
| | | | |
| | + | P2Y12R contains only one <scene name='48/483887/4ntj_disulfide_bond/1'>disulfide bond</scene> that connects the <font color='blue'>amino terminus</font> with <font color='darkorgange'>helix VII</font>. There are also two cholesterol molecules that are bound to two receptor molecules. As displayed in this <scene name='48/483887/Binding/2'>scene</scene>, one cholesterol molecule is bound to a receptor molecule between <font color='deepbluesky'>helix III</font> and <font color='lime'>helix V</font>. Another cholesterol molecule is bound to a receptor molecule shown <scene name='48/483887/Molecule/2'>here</scene> at the interface of <font color='blue'>helix I</font> and <font color='darkorange'>helix VII</font> . |
| | | | |
| - | --
| + | P2Y12R has some distinctive features from other GPCR structures in its family. <font color='lime'>Helix V</font>, for example, has around two more helical turns and does not have the typical helical bend that other GPCR structures have. As mentioned above, <font color='lime'>helix V</font> is <scene name='48/483887/Helix_vii/1'>elongated and straightened</scene> because the structure lacks proline and glycine residues to destabilize its structure. Furthermore, the elongated and straightened conformation causes P2Y12R’s extracellular end to shift 6 Å closer to <font color='turquoise'>helix IV</font> compared to other class A GPCR structures. In addition, the intracellular tip of <font color='darkorange'>helix VII</font> is closer to the seven transmembrane helical bundle. <font color='gold'>Helix VI’s</font> intracellular tip is tilted slightly outward and shifted closer to the intracellular surface than other GPCR structures. |
| | + | |
| | + | This <scene name='48/483887/Polar__nonpolar/1'>view</scene> demonstrates the polar and nonpolar regions of the P2Y12R's structure. AZD1283 spans more than 17 Å between <font color='turquoise'>helix IV</font> and <font color='darkorange'>helix VII</font> contributing to the polar and hydrophobic bonding with helices III–VI. |
| | | | |
| - | ===Overall Structure=== | |
| - | <Structure load='1a84' size='500' frame='true' align='right' caption='This conformation shown above has the widening and flattening of the minor groove of the DNA molecule which resembles A-DNA not found in B DNA. This change is due to the guanine bases that cisplatin interacts with. They compact the major groove and unwind the DNA.' scene='Sandbox_Reserved_430/Intra-strand_phosphate/1' /> | |
| - | The original view shows a double stranded DNA helix and the cisplatin ligand. It is a "duplex dodecamer" d(CCTCTG*G*TCTCCGGAGACCAGAGG), and the asterisks are denoting which base pairs the Ciplatin binds to. This molecule is in its <scene name='Sandbox_Reserved_430/Z-dna_form_due_to_cisplatin/1'>A-DNA conformation due to Cisplatin</scene> which means it stll has the right handed helix, but the widened minor groove distorts its structure. To prove this, the animation shows the distance between intrastrand phosphate groups has been changed due to the insertion of the Cisplatin. Based on the chart found in <font color='purple'>([[Forms of DNA]])</font>, the distance should changed from 7 [Å] to 5.9 [Å]. It is much more rare than the common B-DNA[2]<scene name='Sandbox_Reserved_430/Intra-strand_phosphate/1'>Intra-strand Phosphate</scene> <ref>Takahara, P. M., Rosenzweig, A. C., Frederick, C. A., and Lippard, S. J. (1995) Nature 377, 649-652. Takahara, P. M., Frederick, C. A., and Lippard, S. J. (1996) J. Am. Chem. Soc 118, 12309-12321</ref>. | |
| | | | |
| | | | |
| - | <scene name='Sandbox_Reserved_430/Cisplatin_ligand/1'>Platination bond d(GpG)</scene>. The cisplatin ligand is a cis-diammineplatinum molecule, which is a platinum atom attached to two N7 nitrogen atoms, each apart of a <font color='blue'>guanine bases</font>, and two NH3 molecules attached to the other side. They attach to the 6 and 7 <font color='blue'>guanine bases</font> which links the two bases together and alters the bend in the helix by 49 degrees.The guanine still pair with the 18 and 19 <font color='red'>cytosine bases.</font><ref>Fichtinger-Schepman, A. M. J., van der Veer, J. L., den Hartog, J. H. J., Lohman, P. H. M., and Reedijk, J. (1985) Biochemistry | + | ==Binding Interactions== |
| - | 24, 707-713.</ref>
| + | The interaction between P2Y12 and AZD1283 is different in P2Y12 binding pocket and its PAR1 equivalent. PAR1's 24 residues of ECL2 have more interaction in <scene name='48/483887/Ligand/1'>ligand</scene>binding, while 16 unresolved residues ECL2 of P2Y12 is less likely to interact with AZD1283. Moreover, the shifted outward of helicies IV, VI and VII due to the extracellular make AZD1283 binds deeper into 7TM domain. It formed two pockets for the binding of AZD1283, separated by residues Y105 and K280, with pocket 1 consist of helices III-VII, while pocket 2 consists of helices I-III and VII. Among them, pocket 1 take part in the binding of AZD1283 and P2Y12, while pocket 2 does not. |
| | | | |
| - | Normally, the Cisplatin molecule has two Cl atoms attached to Pt's last two electrons <font color='purple'>[PtCl2(NH3)2]</font>. The Cl atoms can be displaced by "aquation" to form <font color='purple'>[PtCl(H2O)(NH3)2]+</font>. The resulting ligand can be linked to bases now and guanine is the preferred choice. It can then cross-link as in the Cisplatin molecules shown above to form <font color='purple'>[Pt(guanine-DNA)2(NH3)2]+</font>.<ref>http://en.wikipedia.org/wiki/Cisplatin,last accessed 4/8/12.</ref> The N-Pt-N angles, two N7 nitrogens from the guanine base and two NH3 attached directly as part of the Cisplatin ligand, are planar and at 90 degrees, as well as each being a distance of 2.05 [Å] from the PT atom. Another interesting feature about the platinated lesion is in a 5 base section between C4-G21 and T8-A17. The minor groove of the molecule is roughly 9-12 [Å] with a depth range of roughly .4-2.5 [Å] which is much shorter than the B form with a depth of 6.5 [Å]<ref>Andrew Gelasco and Stephen J. Lippard* Department of Chemistry, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139
| + | The binding between P2Y12 and AZD1283 is also different from other GPCRs. The 17A elongated ligand is between helices IV and VII, which belongs in pocket 1. The antagonist AZD1283's piperidinyl-nicotinate group is inserted into sub-pocket of helices III, IV and V; while the benzylsulphonyl group interacts with helices VI and VII, forming at least seven polar and ionic interaction between P2Y12 and AZD1283. |
| - | ReceiVed December 30, 1997; ReVised Manuscript ReceiVed March 27, 1998</ref>.
| + | |
| | | | |
| | + | <scene name='48/483887/C97/1'>C97</scene> and C175 (two cysteine residues in helix III and ECL2 of P2Y12, respectively) are also notable in P2Y12-AZD1283 complexes. The two cysteines are highly conserved in GPCR family, and they form a disulphide bond in all GPCRs. But for P2Y12, no electron density is observed at C97, which means the disulphide bond in P2Y12 would be different. Moreover, mutation at C97 and C175 does not change the protein yield and stability, while increasing the melting temperature when in complex in AZD1283, which indicates that the mutation in both cysteine does not alter P2Y12 binding ability with AZD1283. |
| | | | |
| | | | |
| | | | |
| | + | ==Additional Features== |
| | + | Acute coronary syndrome, a condition in which there is sudden blockage of blood flow to the heart, is majorly caused by a disease known as atherothrombosis. This disease is characterized by blocked arteries due to thrombosis: formation of a clot within a blood vessel. The P2Y12 protein is an important platelet receptor inhibitor that can be combined with aspirin in the management of ACS. It regulates certain functions through purinergic signaling, which is a form of extracellular signaling mediated by purine nucleotides and nucleosides like ADP and ATP.[5] |
| | | | |
| | + | The P2Y12 receptor is mainly found on the surface of blood platelets, and functions as a regulator in platelet activation and blood clotting. The P2Y12 receptor is a G-protein coupled receptor, <scene name='48/483887/Ss/1'>a seven-transmembrane domain protein</scene> (characterized by <font color='pink'>seven alpha helices each</font>) linked to the cAMP-signaling pathway. It mediates platelet activation by decreasing intracellular cAMP levels through inhibition of an AC-mediated signaling pathway.[1] Acting as a chemoreceptor, P2Y12 utilizes ADP as an agonist, which initiates ADP-induced platelet aggregation. Clopidogrel in covalent binding complex with P2Y12 acts as an anti platelet agent, specifically as an ADP receptor inhibitor to decrease platelet aggregation and inhibit thrombus formation.[3] |
| | | | |
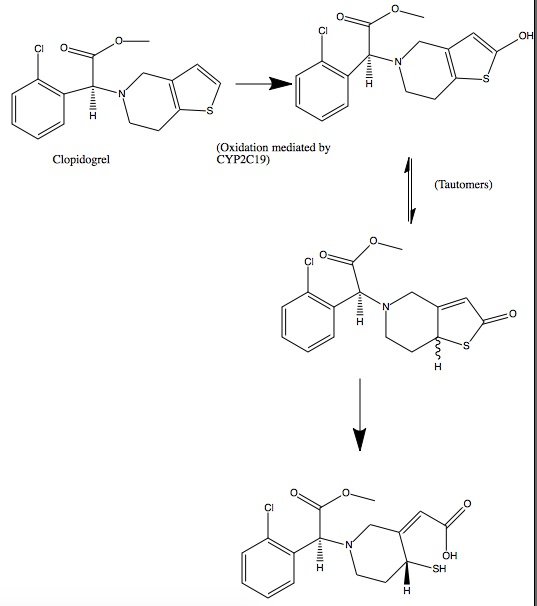
| | + | Clopidogrel is a pro-drug and thienopyridine-type inhibitor of the P2Y12 receptor, which requires Cytochrome P450 to hepatically transform it to exert its anti platelet effect.[4] <scene name='48/483887/Aa/1'>Cytochrome P450</scene> is a membrane protein, characteristic of alternating <font color='gray'>hydrophobic</font> and <font color='pink'>polar</font> groups. The central heme group is stabilized by several side chains. The <font color='orange'>Fe2+</font> atom that makes up the heart of the heme group is surrounded by a <font color='blue'>highly hydrophobic porphyrin ring</font>. The role of the heme group in biological systems is to facilitate oxygen transport as well as aid in electron transfer as part of the electron transport chain. The heme group of the hemeprotein Cytochrome P450 acts as a catalyst for the metabolism of clopidogrel.[2] Shown below is the activation of Clopidogrel in vivo. CYP2C19 is an enzymatic member of the cytochrome P450 mixed-function oxidase system. The final step is a hydrolysis to yield the active metabolite. |
| | | | |
| | + | [[Image:Clop converted.jpg]] |
| | | | |
| | | | |
| | + | ==Quiz Question 1== |
| | + | Below is the structure for AZD1283. <scene name='48/483887/4ntj_azd_surroundingaas/5'>This scene </scene> shows a representation of the pocket where AZD1283 binds to P2Y12R. The dark salmon molecule is AZD1283 which is surrounded by magenta colored amino acids which are polar and gray colored ones which are hydrophobic.List 3 amino acid residues from the scene that should participate in hydrogen bonding with AZD1283 and 3 that should not. Why might the amino acids that don't hydrogen bond with AZD1283 still be present in the pocket? |
| | | | |
| | + | [[Image:AZD Structure.jpg]] |
| | | | |
| - | --
| + | ==See Also== |
| - | ===Binding Interactions=== | + | *[[4ntj]] |
| - | <Structure load='1a84' size='500' frame='true' align='right' caption='Cisplatin' scene='Insert optional scene name here' />
| + | *[[4pxz]] |
| - | As described above, the cisplatin ligand binds to the N7 atoms of the adjacent G6 and G7 guanine bases in a strand of DNA. The green screen
| + | *[[4py0]] |
| - | <scene name='Sandbox_Reserved_430/Binding_site/1'>binding interactions</scene> shows the platinum atom in pink, which is bound to the two N7 atoms of gaunine labeled in green. The N7 atoms are bound to the platinum atom in the ligand, creating a bend in the helix towards the guanine bases of 49 degrees and a total bend in the DNA of 79 degrees<ref>Gelasco, Andrew. "NMR solution and structure of DNA Dodecamer Duplex Containing cis-Diammaineplatium" Department of Chemistry, MIT:1998</ref>. The guanine bases are favored over the adenine because of hydrogen bonding between the amine-hydrogens of the cisplatin and the O=C6 moiety of guanine<ref>Baik MH, Friesner RA, Lippard SJ. "Theoretical Study of Cisplatin binding to purine bases: why doe cisplatin prefer guanine over adenine?" J Am Chem Soc, 2003 Nov 19;125(46):14082-92. 1.↑ 2.0 2.1 2.2 2.3 2.4 2.5 Wang G, Vasquez KM. Z-DNA, an active element in the genome. Front Biosci. 2007 May 1;12:4424-38. PMID:17485386 </ref>. The <scene name='Sandbox_Reserved_430/H-bonding/1'>hydrogen bonding</scene> is shown in this green screen, between the green labeled ammine hydrogens and the oxygen atom labeled in pink. The resulting platination also causes the duplex to unwind by approximately 25 degrees at the site of platination from the base pair T8-A17 to T5-A20. The <scene name='Sandbox_Reserved_430/Minor_groove/6'>minor groove</scene> screen shows the thymine molecules in pink and the adenine ones in green, showing the opening of the helix. These distortions in the duplex allow the minor groove opposite the platinum to be opened to 9.0-12 angstroms, making it shallow and wide.
| + | *[[1vz1]] |
| | | | |
| - | HMG-domain proteins (High-mobility group) bind to recognition sequences found in the minor groove. The bend in DNA caused by cisplatin leaves the minor groove more vulnerable and open for recognition by HMG-domain proteins. Since the expression of these proteins are correlated to tumor cells, the recognition of them by cisplatin-bound DNA could lead to a therapy of cancerous tumors<ref>Gelasco, Andrew. "NMR solution and structure of DNA Dodecamer Duplex Containing cis-Diammaineplatium" Department of Chemistry, MIT:1998</ref>.
| + | ==Credits== |
| | | | |
| | + | Introduction - Adam Murphy |
| | | | |
| | + | Overall Structure - Cora Ricker |
| | | | |
| | + | Drug Binding Site - Duy Nguyen |
| | | | |
| | + | Additional Features - Lauren Timmins |
| | | | |
| | + | Quiz Question 1 - Aidan Finnerty |
| | | | |
| | + | ==References== |
| | + | <references/> |
| | + | 1. Savi, P., et al. "P2Y 12, a new platelet ADP receptor, target of clopidogrel." Biochemical and biophysical research communications 283.2 (2001): 379-383. |
| | | | |
| | + | 2. Coon, Minor J. "Cytochrome P450: nature's most versatile biological catalyst." Annu. Rev. Pharmacol. Toxicol. 45 (2005): 1-25. |
| | | | |
| | + | 3. Gurbel, Paul A., et al. "Clopidogrel for coronary stenting response variability, drug resistance, and the effect of pretreatment platelet reactivity." Circulation 107.23 (2003): 2908-2913. |
| | | | |
| | + | 4. Barn, Kulpreet, and Steven R. Steinhubl. "A brief review of the past and future of platelet P2Y12 antagonist." Coronary artery disease 23.6 (2012): 368-374. |
| | | | |
| - | | + | 5. Bodin, Philippe, and Geoffrey Burnstock. "Purinergic signalling: ATP release." Neurochemical research 26.8-9 (2001): 959-969. |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | --
| + | |
| - | | + | |
| - | ===Additional Features===
| + | |
| - | <Structure load='1a84' size='500' frame='true' align='right' caption='The width of the minor groove of the cisplatin-DNA complex can be compared to the minor groove width of HMG protein-DNA complexes. The similar widths are evidence that Cisplatin is affiliated with HMG proteins.' scene='Insert optional scene name here' />
| + | |
| - | Structural studies of Cisplatin-modified DNA are underway in hope to find a significant correlation
| + | |
| - | between Cisplatin distorted DNA and its ability to bind to high mobility group proteins (HMG proteins).
| + | |
| - | HMG proteins are responsible for many actions within the cell such as transcription, replication and DNA repair.
| + | |
| - | Studies show that over/under expression of these proteins may be the cause of tumors.
| + | |
| - | On the other hand, if HMG proteins attach to a Cisplatin modified double helix, then this may prevent
| + | |
| - | the excision of the helix to be repaired. Resulting in DNA destruction.
| + | |
| - | | + | |
| - | HMG proteins bind to the minor groove of the DNA duplex where their recognition sequences are located.
| + | |
| - | Cisplatin is known to cause the bending of DNA helix as well as the extension of the minor groove width.
| + | |
| - | This opening of the minor groove allows HMG domain proteins to attached to their recognition sequences within
| + | |
| - | the minor groove DNA base pairs
| + | |
| - | <scene name='Sandbox_Reserved_430/Cisplatin_minor_groove_distanc/1'>Cisplatin minor groove</scene>
| + | |
| - | | + | |
| - | This deformation of the DNA duplex with cisplatin forms important complexes with HMG proteins, such as <font color='orange'>LEF-1</font> and hSRY. Binding of these proteins to the already damaged DNA causes further bending. The <font color='orange'>LEF-1</font> HMG protein structure was determined by experiment and superimposed over the known cisplatin-DNA structure.<ref> Gelasco, Andrew. "NMR solution and structure of DNA Dodecamer Duplex Containing cis-Diammaineplatium" Department of Chemistry, MIT:1998</ref> The best fit was shown be over the portion of the cisplatin-DNA structure containing the platinated guanonsines of the 1,2 intrastrand cross link, the similarity holds a good overlap of RMSD of 3.2A.
| + | |
| - | | + | |
| - | <scene name='Sandbox_Reserved_430/2lef/1'>Lef-1 Minor groove</scene>
| + | |
| - | | + | |
| - | This experiment showed that the distortion caused by the <font color='orange'>LEF-1</font> protein is very similar to that caused buy the
| + | |
| - | binding of cisplatin. This comparison of <font color='orange'>LEF-1</font> to cisplatin-modified DNA brings structural evidence that
| + | |
| - | cisplatin –modified DNA may signal the recognition of HMG proteins.
| + | |
| - | | + | |
| - | Another important example is the <font color='red'>HMG1</font> protein binding to the Cisplatin complex. This <font color='red'>HMG1</font>protein is known to bind to the Cisplatin DNA minor groove about the hydrophobic kink created by the distortion. Evidence shows that the phenylalanine residue <font color='red'>HMG1</font> protein is essential for <font color='red'>HMG1</font> interaction with DNA. <ref>Love, JJ. "Structural basis for DNA bending by the architectural transcription factor LEF-1." PubMed:1995 http://www.rcsb.org/pdb/explore/explore.do?structureId=2LEF</ref>Substitution experiments of the phenylalanine with alanine showed that <font color='red'>HMG1</font>HMG1 binding reduced, therefore <font color='red'>HMG1</font> binding is dependent on the phenylalanine and the hydrophobic notch
| + | |
| - | <scene name='Sandbox_Reserved_430/Hmg1_to_cisplatin/6'>HMG1 and the hydrophobic notch</scene>
| + | |
| - | | + | |
| - | <scene name='Sandbox_Reserved_430/Hmg1_to_cisplatin/8'>HMG1-DNA minor groove distance</scene>
| + | |
| - | | + | |
| - | ===Credits===
| + | |
| - | | + | |
| - | Introduction - Gina Lein
| + | |
| - | | + | |
| - | Overall Structure - Greg Keohane
| + | |
| - | | + | |
| - | Drug Binding Site - Louis Pires
| + | |
| - | | + | |
| - | Additional Features - Nicole Hofstetter
| + | |
| - | | + | |
| - | ===References===
| + | |
| - | <references/>
| + | |
|
Introduction
The goal of pharmaceuticals is to prevent or cure disease through drug therapy by specifically targeting cells, proteins, enzymes, genes, etc. It is often crucial to understand the structure, function, and relevant mechanisms involved with the target when designing an effective drug candidate. Furthermore, being able to know the effects on structure after drug-binding can provide insight into the functionality of a specific target. Because of this fact, it is common practice for research labs to develop protein crystals and use methods like x-ray diffraction, electron density mapping, and nuclear magnetic resonance spectroscopy to model the full structure. In this case, P2Y12, a member of P2Y receptor, was modeled when binded to AZD1283, an engineered receptor inhibitor.The molecular scene shows the chemical of P2Y12 with the anionic side chains in red and charged nucleic acids and ligands in grey for contrast.
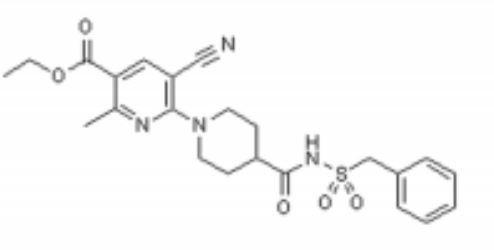
Image analysis of P2Y12 crystals is used to model protein structure in complex with AstraZeneca’s novel AZD1283: Ethyl 6-(4(-((benzylsulphonyl)carbamoyl)piperidin-1yl)-5-cyano-2-methylnicotinate. AZD1283 functions to block the P2Y12 receptor as a means to treat thrombosis. AZD1283-binding leads to unique protein structure, unfound in other P2Y receptors. Helix V of seven transmembrane helices is found to be . This change along with the discovery of a potential second active within P2Y12 has implications on how P2Y12 uses it’s seven transmembrane helical bundle interact with ADP in the bloodstream.
As a member of a large class of G-protein-coupled receptors, P2Y12 is often an initial role player in signal transduction and cellular response due to external environmental factors. In the case of P2Y12, the receptor responds to ADP concentrations in the extracellular matrix and on a larger scale the blood stream. There is importance is understanding how P2Y12's structure receives ADP as an activator. In turn, knowledge of how P2Y12 is affected when properly inhibited can lead to improved drug design in terms of bioavailability, binding affinity, and effectiveness of inhibition. Ultimately, pharmaceuticals will be better able to prevent and treat cardiovascular diseases and medical conditions (thrombosis, hypercoagulable states) and more immediate dangers (stroke, embolism, and heart attacks).
Overall Structure
P2Y12R is a 1 chain structure. The of P2Y12R consists of eight alpha helices. Seven transmembrane alpha helices are tilted and in a bundle, while the carboxy-terminal helix VIII is parallel to the membrane bilayer. The demonstrates how the chain goes from the N to C termini with each helix being approximately one color each of the color scheme.
P2Y12R contains only one that connects the amino terminus with helix VII. There are also two cholesterol molecules that are bound to two receptor molecules. As displayed in this , one cholesterol molecule is bound to a receptor molecule between helix III and helix V. Another cholesterol molecule is bound to a receptor molecule shown at the interface of helix I and helix VII .
P2Y12R has some distinctive features from other GPCR structures in its family. Helix V, for example, has around two more helical turns and does not have the typical helical bend that other GPCR structures have. As mentioned above, helix V is because the structure lacks proline and glycine residues to destabilize its structure. Furthermore, the elongated and straightened conformation causes P2Y12R’s extracellular end to shift 6 Å closer to helix IV compared to other class A GPCR structures. In addition, the intracellular tip of helix VII is closer to the seven transmembrane helical bundle. Helix VI’s intracellular tip is tilted slightly outward and shifted closer to the intracellular surface than other GPCR structures.
This demonstrates the polar and nonpolar regions of the P2Y12R's structure. AZD1283 spans more than 17 Å between helix IV and helix VII contributing to the polar and hydrophobic bonding with helices III–VI.
Binding Interactions
The interaction between P2Y12 and AZD1283 is different in P2Y12 binding pocket and its PAR1 equivalent. PAR1's 24 residues of ECL2 have more interaction in binding, while 16 unresolved residues ECL2 of P2Y12 is less likely to interact with AZD1283. Moreover, the shifted outward of helicies IV, VI and VII due to the extracellular make AZD1283 binds deeper into 7TM domain. It formed two pockets for the binding of AZD1283, separated by residues Y105 and K280, with pocket 1 consist of helices III-VII, while pocket 2 consists of helices I-III and VII. Among them, pocket 1 take part in the binding of AZD1283 and P2Y12, while pocket 2 does not.
The binding between P2Y12 and AZD1283 is also different from other GPCRs. The 17A elongated ligand is between helices IV and VII, which belongs in pocket 1. The antagonist AZD1283's piperidinyl-nicotinate group is inserted into sub-pocket of helices III, IV and V; while the benzylsulphonyl group interacts with helices VI and VII, forming at least seven polar and ionic interaction between P2Y12 and AZD1283.
and C175 (two cysteine residues in helix III and ECL2 of P2Y12, respectively) are also notable in P2Y12-AZD1283 complexes. The two cysteines are highly conserved in GPCR family, and they form a disulphide bond in all GPCRs. But for P2Y12, no electron density is observed at C97, which means the disulphide bond in P2Y12 would be different. Moreover, mutation at C97 and C175 does not change the protein yield and stability, while increasing the melting temperature when in complex in AZD1283, which indicates that the mutation in both cysteine does not alter P2Y12 binding ability with AZD1283.
Additional Features
Acute coronary syndrome, a condition in which there is sudden blockage of blood flow to the heart, is majorly caused by a disease known as atherothrombosis. This disease is characterized by blocked arteries due to thrombosis: formation of a clot within a blood vessel. The P2Y12 protein is an important platelet receptor inhibitor that can be combined with aspirin in the management of ACS. It regulates certain functions through purinergic signaling, which is a form of extracellular signaling mediated by purine nucleotides and nucleosides like ADP and ATP.[5]
The P2Y12 receptor is mainly found on the surface of blood platelets, and functions as a regulator in platelet activation and blood clotting. The P2Y12 receptor is a G-protein coupled receptor, (characterized by seven alpha helices each) linked to the cAMP-signaling pathway. It mediates platelet activation by decreasing intracellular cAMP levels through inhibition of an AC-mediated signaling pathway.[1] Acting as a chemoreceptor, P2Y12 utilizes ADP as an agonist, which initiates ADP-induced platelet aggregation. Clopidogrel in covalent binding complex with P2Y12 acts as an anti platelet agent, specifically as an ADP receptor inhibitor to decrease platelet aggregation and inhibit thrombus formation.[3]
Clopidogrel is a pro-drug and thienopyridine-type inhibitor of the P2Y12 receptor, which requires Cytochrome P450 to hepatically transform it to exert its anti platelet effect.[4] is a membrane protein, characteristic of alternating hydrophobic and polar groups. The central heme group is stabilized by several side chains. The Fe2+ atom that makes up the heart of the heme group is surrounded by a highly hydrophobic porphyrin ring. The role of the heme group in biological systems is to facilitate oxygen transport as well as aid in electron transfer as part of the electron transport chain. The heme group of the hemeprotein Cytochrome P450 acts as a catalyst for the metabolism of clopidogrel.[2] Shown below is the activation of Clopidogrel in vivo. CYP2C19 is an enzymatic member of the cytochrome P450 mixed-function oxidase system. The final step is a hydrolysis to yield the active metabolite.
Quiz Question 1
Below is the structure for AZD1283. shows a representation of the pocket where AZD1283 binds to P2Y12R. The dark salmon molecule is AZD1283 which is surrounded by magenta colored amino acids which are polar and gray colored ones which are hydrophobic.List 3 amino acid residues from the scene that should participate in hydrogen bonding with AZD1283 and 3 that should not. Why might the amino acids that don't hydrogen bond with AZD1283 still be present in the pocket?
See Also
Credits
Introduction - Adam Murphy
Overall Structure - Cora Ricker
Drug Binding Site - Duy Nguyen
Additional Features - Lauren Timmins
Quiz Question 1 - Aidan Finnerty
References
- ↑ Zhang K, Zhang J, Gao ZG, Zhang D, Zhu L, Han GW, Moss SM, Paoletta S, Kiselev E, Lu W, Fenalti G, Zhang W, Muller CE, Yang H, Jiang H, Cherezov V, Katritch V, Jacobson KA, Stevens RC, Wu B, Zhao Q. Structure of the human P2Y12 receptor in complex with an antithrombotic drug. Nature. 2014 May 1;509(7498):115-8. doi: 10.1038/nature13083. Epub 2014 Mar 23. PMID:24670650 doi:http://dx.doi.org/10.1038/nature13083
1. Savi, P., et al. "P2Y 12, a new platelet ADP receptor, target of clopidogrel." Biochemical and biophysical research communications 283.2 (2001): 379-383.
2. Coon, Minor J. "Cytochrome P450: nature's most versatile biological catalyst." Annu. Rev. Pharmacol. Toxicol. 45 (2005): 1-25.
3. Gurbel, Paul A., et al. "Clopidogrel for coronary stenting response variability, drug resistance, and the effect of pretreatment platelet reactivity." Circulation 107.23 (2003): 2908-2913.
4. Barn, Kulpreet, and Steven R. Steinhubl. "A brief review of the past and future of platelet P2Y12 antagonist." Coronary artery disease 23.6 (2012): 368-374.
5. Bodin, Philippe, and Geoffrey Burnstock. "Purinergic signalling: ATP release." Neurochemical research 26.8-9 (2001): 959-969.
|


