Sandbox Reserved 431
From Proteopedia
(→Additional Features) |
|||
| (287 intermediate revisions not shown.) | |||
| Line 3: | Line 3: | ||
<!-- INSERT YOUR SCENES AND TEXT BELOW THIS LINE --> | <!-- INSERT YOUR SCENES AND TEXT BELOW THIS LINE --> | ||
| - | ==''' | + | =='''Vitamin D activation by cytochrome P450, Rickets (3c6g)<ref>PMID: 18511070 </ref>'''== |
| - | + | by Isabel Hand, Elizabeth Humble, Kati Johnson, Samantha Kriksceonaitis, and Matthew Tiller | |
| - | + | [[Student Projects for UMass Chemistry 423 Spring 2016]] | |
| - | + | ||
| - | + | ==Introduction== | |
| + | Rickets is a disease resulting from prolonged vitamin D deficiency. As vitamin D is vital for the absorption of phosphorus and calcium, a deficiency would cause weakening of bones. Rickets also causes muscle weakness, skeletal deformities, dental problems, inhibition of growth, and muscle spasms. In some cases, rickets can be inherited due to mutations in genes responsible for coding for human CYP2R1. | ||
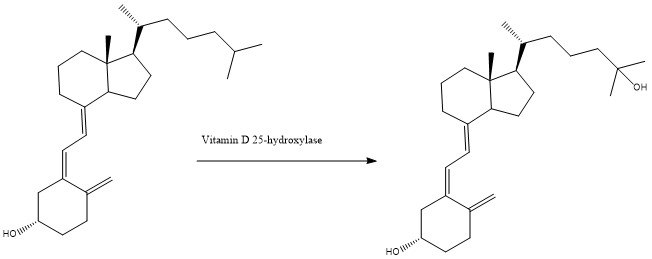
| - | + | In the human body, CYP2R1, a member of the cytochrome P450 family, is responsible for the first steps of the conversion of vitamin D into a bioavailable form within the liver. CYP2R1 is also known as Vitamin D 25-hydroxylase as it hydroxylates vitamin D3 into calcidiol, the bioavailable form of the vitamin, which would then be converted to calcitriol via the enzyme 25-hydroxyvitamin D3 1-alpha-hydroxylase, as seen in figure 1. | |
| - | + | [[Image:Action_Of_CYP2R1.jpg]] | |
| - | + | Fig. 1, the conversion of vitamin D3 into calcidiol via Vitamin D 25-hydroxylase | |
| - | + | ||
| + | <scene name='48/483888/Human_p450/1'>Human P450 Cytochrome</scene> is shown with amino acids in teal and the heme center shown as a space filling model, with the nitrogen shown in blue, carbon in gray, oxygen in red, and the iron center shown in orange | ||
| + | <StructureSection load='3c6g' size='350' side='right' caption='Structure of Human Cytochrome p450' scene=''> | ||
| + | ==Overall Structure== | ||
| + | Cytochrome P450 is an <scene name='48/483888/Cytochrome_p450_dimer/1'>asymmetric dimer</scene>, which means the protein is made of two subunits that are structurally very similar to one another, but not identical. Each dimeric subunit contain 12 α-helices (labeled A-L) along with some β-sheets that are localized to one side of the molecule. Helices F and G from each of the units form the dimer interface of cytochrome P450, and are involved in the formation of the active site. This dimeric interface of the protein is stabilized by <scene name='48/483888/Cytochrome_p450_interface/1'>hydrogen bonding interactions</scene> between residues from the G helix of one the units with residues located on the the F helix of the second unit, and vice versa. Two molecules of 2-hydroxypropyl-β-cyclodextrin are also found near the dimer interface. The cyclodextrins are believed to help further stabilize the protein, and also shield the hydrophobic part of the F-G helix transition loop from the solvent by <scene name='48/483888/Cytochrome_p450_cyclodextrins/1'>encapsulating the Phe240 residue within its cavity</scene>. Cytochrome P450 has an apparent mass of ~120 kDa. | ||
| + | ==Binding Interactions== | ||
| + | CYP2R1 binds vitamin D3 at an extended binding site that orients the bound molecule to bring its side chain close to the heme and allow for hydroxylation. The binding site is located at the channel between the G and I helices and the B' helix/B-C loop. The active site has non-polar residues which allows for nonpolar interactions with D3. In the <scene name='48/483888/Spacefilled_binding_site/1'>space filling representation </scene> you can see the residues that interact to bind Vitamin D3 and the channel between. Once bound, the D3 molecule is submerged into the protein, with only its 3-OH group showing. | ||
| + | The B' helix is one of the substrate recognition sites and has a flexible C terminus which unwinds outward to allow entrance of the substrate into the active site channel. Due to the stabilizing interactions of B' with the F-G loop, binding of the substrate causes the protein to adopt a closed conformation which closes the access channel. | ||
| - | <Structure load='1nny' size='500' frame='true' align='left' caption='PTP1B catalytic loop' scene='Insert optional scene name here' /> | ||
| - | <br><br> | ||
| - | ===Overall Structure=== | ||
| - | + | ==Additional Features== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Cytochrome P450 has a central iron-bound heme, which, combined with its structural conformation, allows for hydroxylation with the attached substrate. Cytochrome P450 has specific vitamin D 25-hydroxylase activity, which does not function properly when a person has rickets. Rickets is caused by a lack of sufficient vitamin D in their system, which is often caused by a vitamin D-25 hydroxylation defect. Leu99Pro is an evolutionarily conserved mutation in the beta helix which contributes to the hydroxylation defect. Leu99 does not inhibit substrate binding; however, Leu99Pro disturbs hydrogen binding around the heme and interferes with the helix steric properties, causing protein instability. When Leu99 does not have the proline mutation, its carboxyl group forms hydrogen bonds with Arg445, which are both located around the central heme and allows for hydroxylation of vitamin D. | |
| - | PTP1B is thought to primarily be responsible for the dephosphorylation of the insulin receptor and, therefore, acts to downregulate insulin signaling. Inhibiting PTP1B is linked to improved insulin response and activation of the insulin pathway. PTP1B deficient mice showed a resistance to diet induced diabetes. | ||
| - | <Structure load='1nny' size='500' frame='true' align='right' caption='Enzyme active site' scene='Insert optional scene name here' /> | ||
| - | Overall the <scene name='Sandbox_Reserved_431/Working_ligand/1'>ligand</scene> is a competitive inhibitor which reversibly binds to two sites simultaneously and has a high selectivity for PTP1B over other phosphatases. This ligand was engineered by a research group from Abbot labs and the steps they took in its design revolve around many important binding interactions. The group designed the drug in <scene name='Sandbox_Reserved_431/Ligand_design/1'>three different stages</scene>. In order to identify this inhibiting ligand, the research group of interest first used a computer to scan a library of 10,000 organic compounds based on there NMR spectra, searching for those who have an affinity for the enzymes active site. The best match produced by the screen was diaryloxamic acid which the group then modified to <font color = 'dodgerblue'>naphthyloxamic acid</font> , reasoning that an inhibitor that took up more space would be a more potent inhibitor. When the group mixed this molecule with enzyme, NMR data showed a chemical shift in <scene name='Sandbox_Reserved_431/1st_binding/4'>Val49, Gly220, and Gly 218</scene>, part of the active site of the enzyme. The observed kinetics indicated competitive and reversible inhibition. | ||
| - | After their initial success, the group then reasoned that since the active domain of PTP1B is somewhat conserved by many other phosphatases and extremely similar in some ( TCPTP), incorporation of binding to a second and nonactive site would dramatically increase selectivity. Based off of naphthyloxamic acid, the group then created what they called <font color = 'darkorange'>compound 12</font> hoping that it would satisfy these conditions. Mixing the new ligand and enzyme then screening with NMR N-15 and C-13 resonance yielded data showing that in addition to binding to Val49, Gly220, and Gly 218, the new ligand bound to additional active sites, those being <scene name='Sandbox_Reserved_431/Active_site_2/5'>Gln262 and Arg221</scene> The new ligand also forms hydrogen bonds to the backbone of residues <font color = 'darkorange'> Ser216-Gly220</font> as well as forming a hydrophobic interaction with Tyr46. The group improved the site two ligand creating <font color = 'magenta'>compound 23</font> compound 23, which primarily adds interactions with <scene name='Sandbox_Reserved_431/Active_site_2/6'>Met258, Arg24, and Arg254</scene>. It is not yet know whether <font color = 'magenta'> Arg 24, Arg 254,</font> or both residues are interacting with the ligand, but it is believed that they are either forming hydrogen bonds to the ligand or forming a salt bridge. The groups final ligand shows excellent potency for PTP1B while also showing high seletivity against other phosphatases. | ||
| - | + | <scene name='48/483888/Hemegroup/1'>Leu99 and Arg445</scene> | |
| - | < | + | </StructureSection> |
| - | + | ==Quiz Question 1== | |
| - | + | 1A) Why is Proline so poorly suited for inclusion in Alpha Helices? | |
| + | A) It cannot hydrogen bond due to the position of its amide | ||
| + | B) The residue is incapable of forming the correct phi and psi angles in a helix | ||
| + | C) The steric hindrance of its sidechain | ||
| + | D) A and C | ||
| + | E) All of the above | ||
| - | = | + | 1B) This protein being a dimer, has symmetry between its two large sections, from <scene name='48/483888/Gandfhelices/1'>this orientation</scene> where most of the molecule has been cut away for simplicity, you can see where one half (in green) comes within close proximity of the other half (in blue). These 2 pairs of helices help hold the dimer together via electrostatic interactions. If the black residue is Arg, and the white residue is Asp, what is most likely to be on the opposite helix |
| + | (Arg match, Asp match) | ||
| + | A) Asp, His | ||
| + | B) Gly, Val | ||
| + | C) Met, Lys | ||
| - | + | ==See Also== | |
| + | *[[Cytochrome P450]] | ||
| + | *[[Drug Metabolism by CYP450 Enzymes]] | ||
| + | *[[3w0y]] | ||
| + | *[[3w0c]] | ||
| - | + | ==Credits== | |
| - | + | Introduction - Sami Kriksceonaitis | |
| - | + | Overall Structure - Kati Johnson | |
| - | + | Binding Interactions - Isabel Hand | |
| + | Additional Features - Elizabeth Humble | ||
| + | |||
| + | Quiz Question 1 - Matthew Tiller | ||
| + | |||
| + | ==References== | ||
<references/> | <references/> | ||
| - | 3. Edwards, K., T. Davis, D. Marcey, J. Kurihara, D. Yamamoto. 2001. Comparative Analysis of the Band 4.1/ezrin-related Protein Tyrosine Phosphatase Pez from Two Drosophila Species: Implication for Structure and Function. Gene 275: 195-205. | ||
Current revision
| This Sandbox is Reserved from January 19, 2016, through August 31, 2016 for use for Proteopedia Team Projects by the class Chemistry 423 Biochemistry for Chemists taught by Lynmarie K Thompson at University of Massachusetts Amherst, USA. This reservation includes Sandbox Reserved 425 through Sandbox Reserved 439. |
Contents |
Vitamin D activation by cytochrome P450, Rickets (3c6g)[1]
by Isabel Hand, Elizabeth Humble, Kati Johnson, Samantha Kriksceonaitis, and Matthew Tiller
Student Projects for UMass Chemistry 423 Spring 2016
Introduction
Rickets is a disease resulting from prolonged vitamin D deficiency. As vitamin D is vital for the absorption of phosphorus and calcium, a deficiency would cause weakening of bones. Rickets also causes muscle weakness, skeletal deformities, dental problems, inhibition of growth, and muscle spasms. In some cases, rickets can be inherited due to mutations in genes responsible for coding for human CYP2R1.
In the human body, CYP2R1, a member of the cytochrome P450 family, is responsible for the first steps of the conversion of vitamin D into a bioavailable form within the liver. CYP2R1 is also known as Vitamin D 25-hydroxylase as it hydroxylates vitamin D3 into calcidiol, the bioavailable form of the vitamin, which would then be converted to calcitriol via the enzyme 25-hydroxyvitamin D3 1-alpha-hydroxylase, as seen in figure 1.
Fig. 1, the conversion of vitamin D3 into calcidiol via Vitamin D 25-hydroxylase
is shown with amino acids in teal and the heme center shown as a space filling model, with the nitrogen shown in blue, carbon in gray, oxygen in red, and the iron center shown in orange
| |||||||||||
Quiz Question 1
1A) Why is Proline so poorly suited for inclusion in Alpha Helices?
A) It cannot hydrogen bond due to the position of its amide
B) The residue is incapable of forming the correct phi and psi angles in a helix
C) The steric hindrance of its sidechain
D) A and C
E) All of the above
1B) This protein being a dimer, has symmetry between its two large sections, from where most of the molecule has been cut away for simplicity, you can see where one half (in green) comes within close proximity of the other half (in blue). These 2 pairs of helices help hold the dimer together via electrostatic interactions. If the black residue is Arg, and the white residue is Asp, what is most likely to be on the opposite helix
(Arg match, Asp match)
A) Asp, His
B) Gly, Val
C) Met, Lys
See Also
Credits
Introduction - Sami Kriksceonaitis
Overall Structure - Kati Johnson
Binding Interactions - Isabel Hand
Additional Features - Elizabeth Humble
Quiz Question 1 - Matthew Tiller
References
- ↑ Strushkevich N, Usanov SA, Plotnikov AN, Jones G, Park HW. Structural analysis of CYP2R1 in complex with vitamin D3. J Mol Biol. 2008 Jun 27;380(1):95-106. Epub 2008 Apr 8. PMID:18511070 doi:10.1016/j.jmb.2008.03.065