Sandbox Reserved 434
From Proteopedia
(Difference between revisions)
(→Introduction) |
|||
| (282 intermediate revisions not shown.) | |||
| Line 3: | Line 3: | ||
<!-- INSERT YOUR SCENES AND TEXT BELOW THIS LINE --> | <!-- INSERT YOUR SCENES AND TEXT BELOW THIS LINE --> | ||
| - | ==''' | + | =='''Pantetheinase (4CYG)<ref>PMID: 25478849 </ref>'''== |
| - | + | by [Luke Schnitzler, Patrick Tonne, Owen O'Connor, Tyler Russell, Nicholas Sant] | |
| - | + | [[Student Projects for UMass Chemistry 423 Spring 2016]] | |
| - | + | <StructureSection load='4CYG' size='350' side='right' caption='caption for Molecular Playground (PDB entry [[4CYG]])' scene=''> | |
| - | < | + | |
| - | + | ||
| - | + | ||
| - | == | + | ==Introduction== |
| - | < | + | Vanin 1, otherwise known as pantetheinase, is an enzyme found throughout the body in various tissues including the liver and kidneys. As an ectoenzyme—any enzyme found on the outside or outer surface of a cell—pantethiense is anchored to the cell wall by a glycosylphosphatidylinositol (GPI) linker, allowing for the it to carry out its enzymatic purpose of hydrolyzing pantetheine to pantothenic acid and cysteamine [1]. The two protein subunits possess dense regions of <scene name='48/483891/Secondary_structure/1'>beta strands and alpha helices</scene> which create binding sites for three types of ligands, the non-polar <scene name='48/483891/Rrv/1'>RRV ligand</scene>, a di(hydroxyethyl)ether compound <scene name='48/483891/Peg/1'>PEG</scene>, and <scene name='48/483891/Nag/1'>NAG</scene> (N-acetyl-d-glucosamine) ligands. The identification of the associated binding sites has led to the investigation of active-site inhibitors (see Additional Features). |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | The importance of vanin 1 lies in the products of the enzymatic reaction. Pantothenic acid (vitamin B12) plays a significant role in the maintenance of the nervous system and brain. The compound is also involved in DNA synthesis, as well as fatty acid and amino acid metabolism [2]. Cysteamine—a product of the degradation of the amino acid cysteine—is used to form coenzyme A, a compound that plays a key role in the citric acid cycle and the synthesis of fatty acids. A lack of these biomolecules can lead to significant physiological issues (see Additional Features). | |
| - | + | ||
| - | + | ||
| - | Leadzyme requires Pb2+ to cleave RNA and is inhibited by Mg2+, through competitive inhibition. A possible suggestion for Mg2+ inhibition may have to do with the Mg(H2O)6 which bind in the major groove at the G39, G30 and A29 nucleotides at one location (<scene name='Sandbox_Reserved_434/Site_i/2'>Site I</scene>, in both the ground state and pre-catalytic state), at the U47, G20, and G19 nucleotides from another location (<scene name='Sandbox_Reserved_434/Site_iii/3'>Site III</scene>, from both states), and at the U41, G42, G43, G24, and A25 nucleotides in the ground state (<scene name='Sandbox_Reserved_434/U41_g42_g43_g24_and_a25/3'>Site II</scene>). This last interaction is particularly important since it “ligates” the <scene name='Sandbox_Reserved_434/Trinucleotide_bulge/4'>Trinucleotide Bulge</scene>, which inhibits the activity of the leadzyme. Since this type of interaction occurs only in the ground state, it is thought to stabilize the ground state and thus prevent catalysis of RNA cleavage. The Mg2+ "pulls in" the trinucleotide bulge by hydrogen bonding with<font color = 'red'>N7 and O6 in G42 and O4 in U41</font> with a "sphere" of six water molecules attached to the magnesium ion. It is worth noting that because of the competition between metal ions in leadzyme, if lead is added to the ground state form of leadzyme with bound Mg2+ at site II, the structure may change from ground state to pre-catalytic by interrupting the hydrogen bonding/bridging between the two tandem purines (G42 & G43) and A25 and G24, thus relaxing the trinucleotide bulge.There are some differences in the types of binding involved in sites II than in sites I & III. For example. In site II, O6 at the G43 nucleotide shares an H2O with O6 of G42 rather than hydrogen bonding with a separate water molecule. | ||
| - | + | ==Overall Structure== | |
| + | The protein 4CYG has a two chain structure. The chains have identical sequences, consisting of 506 total residues each. There are 87 residues missing from the model, which have been replace with spherical basket-like structures. The structure of only Chain A has been isolated <scene name='48/483891/Chain_a_rainbow/4'>here</scene> in order to provide a more simple view. Chain A is colored to show the sequence direction <scene name='48/483891/Chain_a_rainbow/5'>here</scene> | ||
| - | + | The secondary structure of 4CYG is made up of both beta sheets and alpha helices. The alpha helices of one chain, of which there are 11, are isolated <scene name='48/483891/Alphas_only/2'>here</scene>, where the polar side of the helix is pink and the hydrophobic side is gray. The beta sheets of one chain, colored by polarity, can be viewed <scene name='48/483891/Betas_only_polar/1'>here</scene>. The polar portions are colored pink and nonpolar portions are colored grey, allowing for a visual representation of the alternating sequence. | |
| - | + | 4CYG has three types ligands involved in the structure. There are two <scene name='48/483891/Rrv/1'>RRV</scene> ((2r)-2,4-dihydroxy-n-[(3s)-3-hydroxy-4-phenylbutyl]-3,3-dimethylbutanamide) ligands, two <scene name='48/483891/Peg/1'>PEG</scene> (di(hydroxyethyl) ether) ligands, and eight <scene name='48/483891/Nag/1'>NAG</scene> (N-acetyl-d-glucosamine) ligands. | |
| - | < | + | ==Binding Interactions== |
| - | + | 4CYG is a key protein that is involved in the breakdown of pantetheine to panthothenic acid and cysteamine. These proteins are associated with many metabolic diseases like type 2 diabetes. Understanding the binding interaction would give insight into treating these diseases more effectively. 4CYG has three <scene name='48/483891/Binding_site/1'>Catalytic Residues</scene> (Glu79, Lys178 and Cys211) that represents the active site of the enzyme. The purple amino acids represent the three amino acids directly involved in the binding interactions. The active site is located in the center of the enzyme in between the two sub-units. It was discovered that Glu79 and Lys178 were responsible for orienting and activating Cys211 to catalyze the reaction. The substrate forms a covalent bond with Cys211 producing the transition state.[1] | |
| - | The | + | In addition, the two residues <scene name='48/483891/Glu249_and_glu439/2'>GLU249 AND GLU439</scene> are essential for enzymatic function too. The purple regions are polar whereas the grey regions are hydrophobic. The two black amino acids represent GLU249 and GLU439. The two glutamic acid residues are both located in hydrophobic regions 4 Angstroms away. Although this is energetically unfavorable to have a polar amino acid in a nonpolar region, these two amino acids help maintain the structure between the two sub-units for proper binding interactions to occur.[1] |
| - | + | ==Additional Features== | |
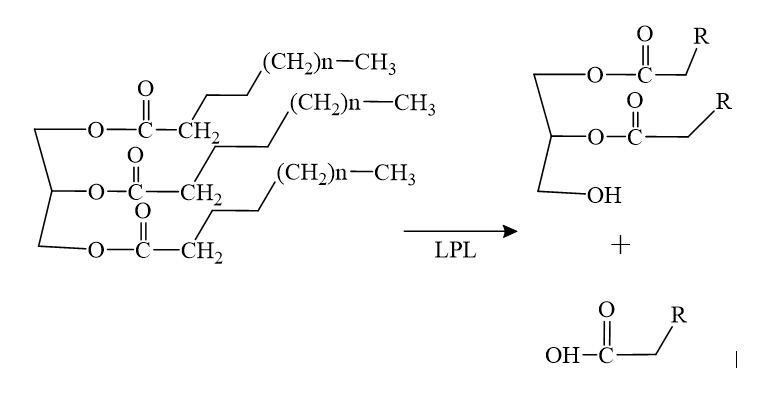
| + | The biological importance of pantetheinase is found in the products cysteamine and vitamin B5, formed from the hydrolysis reaction shown below. Cysteamine and vitamin B5 are key components in the synthesis of other necessary bio-molecules such as acetylcholine and coenzyme A. | ||
| - | The | + | [[Image:rxn.png]] |
| + | ===Inhibition=== | ||
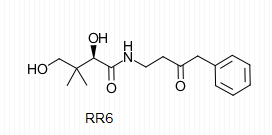
| + | The importance of pantetheinase stems from the vitality of the components of the reaction it catalyzes. It is for this reason that pantetheinase has been a promising point of research in the field of medicine. Pantetheine analogues known as pantothenamides have been shown to act as effective antibiotics that protect the body from bacterial intruders. The similarity of these analogues to pantetheine allows for the active sites on pantetheinase to catalyze their breakdown through hydrolysis. The administration of RR6 in the presence of pantetheinase and other antibiotic pantothenamides revealed that RR6 acts as a competitive inhibitor with great affinity for the active site Cys at <scene name='48/483891/Rr6/2'>residue 211</scene> on pantetheinase, thus preserving desired concentrations of the pantothenamides with antibiotic characteristics. The RR6 inhibitor is shown below. | ||
| - | + | [[Image:RR6.png]] | |
| + | ===Absence=== | ||
| + | Vitamin B5 is the recycled product of the hydrolysis of pantetheine and is a major substrate in the formation of coenzyme A (CoA). Without pantetheinase, the hydrolysis of pantetheine would occur at a significantly slower rate and would therefore produce vitamin B5 and cysteamine at a slower rate. Lower concentrations of Vitamin B5 would result in inadequate prodution of CoA and therefore inadequate production of acetylcholine. Acetylcholine is a neurotransmitter that is needed for proper brain function which means an absence of pantetheinase could lead to neurological complications. | ||
| + | |||
| + | Cysteamine is important in the regulation of cystine levels in lysosomes. A lack of cysteamine due to the absence of pantetheinase would lead to a build up of lysosomal cystine, a disease known as cystinosis. | ||
| - | The <scene name='Sandbox_Reserved_434/Ph_1/5'>duplex</scene> shows residues with <font color='red'>pKa < 3.1</font>, <font color='light blue'>pKa of 4-5</font> and <font color=00007C >pKa > 5</font>. The pH closest to the biological range, residue A45, is an adenine that participates in coordination of the lead at one of three identified lead binding sites. This adenine is also part of the only non-Watson-Crick pair near the active site. The loop connecting the two strands consists of adenines with pKa around 3.5.<ref> Legault, P., Pardi, A. (1997) ''J. Am. Chem. Soc. 119'', 6621-6628</ref> The <scene name='Sandbox_Reserved_434/Ph_2/3'>asymmetrical bulge</scene> does not exhibit an considerable increase in stability at lower pH as experienced by some similar sequences,<ref>PMID: 19485416 </ref> which would be consistent with the pH instead changing the lead binding capability. | ||
| - | == | + | ==Quiz Question 1== |
| - | + | Pantetheinase exhibits an extremely similar sequence to the other proteins in the Vanin family. In addition, <scene name='48/483891/Pantetheinase/1'>pantetheinase</scene> is also sequentially similar to this enzyme, which allows the body to utilize a certain vitamin: | |
| - | + | a. gastric lipase | |
| - | + | b. biotinidase | |
| - | + | c. carboxypeptidase | |
| - | === | + | d. pepsin |
| + | |||
| + | |||
| + | |||
| + | ==See Also== | ||
| + | *[[1h1t]] | ||
| + | *[[1mop]] | ||
| + | *[[1esm]] | ||
| + | |||
| + | ==Credits== | ||
| + | |||
| + | Introduction - Patrick Tonne | ||
| + | |||
| + | Overall Structure - Luke Schnitzler | ||
| + | |||
| + | Drug Binding Site - Owen O'Connor | ||
| + | |||
| + | Additional Features - Nick Saint | ||
| + | |||
| + | Quiz Question 1 - Tyler Russell | ||
| + | |||
| + | ==References== | ||
<references/> | <references/> | ||
| + | [2] "Pantetheinase." VNN1. UniProt, n.d. Web. 10 Apr. 2016. | ||
| + | |||
| + | [3]Jansen, Patrick, and Joost Schalkwijk. "Chemical Biology Tools to Study Pantetheinases of the Vanin Family." Biochemical Society Transactions. Portland Press, n.d. Web. 10 Apr. 2016. | ||
| + | |||
| + | [4]"Cystinosis." Genetics Home Reference. U.S. National Library of Medicine, 8 Apr. 2016. Web. 10 Apr. 2016. | ||
Current revision
| This Sandbox is Reserved from January 19, 2016, through August 31, 2016 for use for Proteopedia Team Projects by the class Chemistry 423 Biochemistry for Chemists taught by Lynmarie K Thompson at University of Massachusetts Amherst, USA. This reservation includes Sandbox Reserved 425 through Sandbox Reserved 439. |
Pantetheinase (4CYG)[1]
by [Luke Schnitzler, Patrick Tonne, Owen O'Connor, Tyler Russell, Nicholas Sant]
Student Projects for UMass Chemistry 423 Spring 2016
| |||||||||||