We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Anum-II
From Proteopedia
(Difference between revisions)
(New page: {{STRUCTURE_1mg6 | PDB=1mg6 | SCENE= }} == '''Introduction''' == Anum-II is a Lys49 Phospholipase A2 (<scene name='Sandbox136/Lys/1'>K49</scene>PLA2),protein extracted from [http...) |
|||
| (2 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <StructureSection load="1mg6" size="400" color="" frame="true" spin="on" Scene= side="right" caption= > | |
| - | + | ||
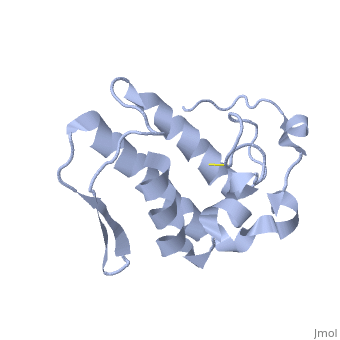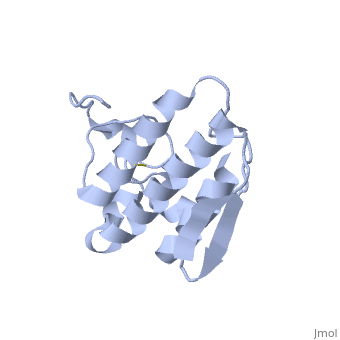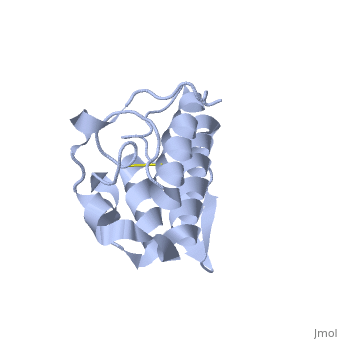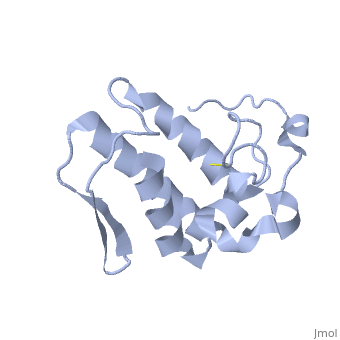
== '''Introduction''' == | == '''Introduction''' == | ||
| Line 9: | Line 8: | ||
| - | Anum-II has a length of 134 amino-acids. The phospholipase is formed by a short N-terminalαhelix (between residues 2-12), a 2nd αhelix (residues 40-55), two-stranded antiparallel <scene name='Sandbox136/Feuillets/1'>sheet</scene> linked thanks to a βwing (74-85) and a 3rd αhelix (residues 90-107). The 3rd αhelix is bound to the 2nd αhelix (in an antiparallele way) thanks to <scene name='Sandbox136/Disulfide/1'>disulfide bonds </scene> ([Cys 44-Cys 105] and [Cys 51-Cys 98]) and thus form a <scene name='Sandbox136/Stabilisation/1'>rigid platform</scene>. The protein is stabilized by 5 other disulfides bonds [Cys 27-Cys 125], [Cys 29-Cys 45] [Cys 50 Cys 134] [Cys 61-Cys 91] [Cys84-Cys96]. Alignment of Anum-II with other PLA2 have revealed that the positions of amino-acid residues which form the <scene name='Sandbox136/Site_catalytique/2'>catalityc apparatus</scene> are conserved (His48,Tyr52, Tyr73 and Asp99) except for Asp49 which is replaced by Lys 49. | + | Anum-II has a length of 134 amino-acids. The phospholipase is formed by a short N-terminalαhelix (between residues 2-12), a 2nd αhelix (residues 40-55), two-stranded antiparallel <scene name='Sandbox136/Feuillets/1'>sheet</scene> linked thanks to a βwing (74-85) and a 3rd αhelix (residues 90-107). The 3rd αhelix is bound to the 2nd αhelix (in an antiparallele way) thanks to <scene name='Sandbox136/Disulfide/1'>disulfide bonds </scene> ([Cys 44-Cys 105] and [Cys 51-Cys 98]) and thus form a <scene name='Sandbox136/Stabilisation/1'>rigid platform</scene>. The protein is stabilized by 5 other disulfides bonds [Cys 27-Cys 125], [Cys 29-Cys 45] [Cys 50 Cys 134] [Cys 61-Cys 91] [Cys84-Cys96]. Alignment of Anum-II with other PLA2 have revealed that the positions of amino-acid residues which form the <scene name='Sandbox136/Site_catalytique/2'>catalityc apparatus</scene> are conserved (<scene name='49/497100/Active_site_residues_with_fas/2'>His48,Tyr52, Tyr73 and Asp99</scene>) except for Asp49 which is replaced by Lys 49. |
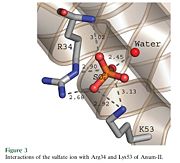
The structure of the protein has revealed the presence of an anion-binding site (Murakami ''et al.'', 2006) between <scene name='Sandbox136/Anion/2'>R34</scene>, <scene name='Sandbox136/Anion/2'>K53</scene> and a water molecule (see Figure 3). Sulfate ion is anchored thanks to hydrogen bonds between: | The structure of the protein has revealed the presence of an anion-binding site (Murakami ''et al.'', 2006) between <scene name='Sandbox136/Anion/2'>R34</scene>, <scene name='Sandbox136/Anion/2'>K53</scene> and a water molecule (see Figure 3). Sulfate ion is anchored thanks to hydrogen bonds between: | ||
| Line 35: | Line 34: | ||
Vascular endothelial growth factor (VEGF165) and its Kinase insert domain containing receptor (KDR) are used in mechanisms of regulations of blood vessel formations. Experiments have revealed that the venom extracted from ''Agkistrodon piscivorus'' contains a KDR binding protein.It has been shown that this KDRb-p was in fact a Lys49 PLA2.Lys49 is able to bind to the extracellular domain of KDR receptors with a sub-nanomolar affinity (Yamazaki ''et al.'', 2005). Studies revealed the fact that the region of the protein involved in the fixation on KDR was located in the C-terminal region (Chioato ''et al.'', 2002). C- terminal region differs from the rest of the protein in his amino acid sequence, indeed some amino-acids are positive (K,R) and seem to play a role in the recognition and fixation on KDR. | Vascular endothelial growth factor (VEGF165) and its Kinase insert domain containing receptor (KDR) are used in mechanisms of regulations of blood vessel formations. Experiments have revealed that the venom extracted from ''Agkistrodon piscivorus'' contains a KDR binding protein.It has been shown that this KDRb-p was in fact a Lys49 PLA2.Lys49 is able to bind to the extracellular domain of KDR receptors with a sub-nanomolar affinity (Yamazaki ''et al.'', 2005). Studies revealed the fact that the region of the protein involved in the fixation on KDR was located in the C-terminal region (Chioato ''et al.'', 2002). C- terminal region differs from the rest of the protein in his amino acid sequence, indeed some amino-acids are positive (K,R) and seem to play a role in the recognition and fixation on KDR. | ||
| - | + | </StructureSection> | |
== Bibliography == | == Bibliography == | ||
| Line 45: | Line 44: | ||
Yamazaki, Y., Matsunaga, Y., Nakano, Y. & Morita, T. (2005). ''J. Biol. Chem''. '''280''', 29989–29992. | Yamazaki, Y., Matsunaga, Y., Nakano, Y. & Morita, T. (2005). ''J. Biol. Chem''. '''280''', 29989–29992. | ||
| + | <br /> | ||
| + | <br /> | ||
| + | Created with the participation of [[User:Elie Peillon|Elie Peillon]] | ||
| + | |||
| + | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
Bibliography
Holland, D.R., Clancy, L.L., Muchmore, S.W., Ryde, T.J., Einspahr, H.M., Finzel, B.C., Heinrikson, R.L., Watenpaugh, K.D. (1990).Biochem-J .265,17649-56
Chioato, L., De Oliveira, A. H., Ruller, R., Sa., J. M., Ward, R. J. (2002).Biochem-J .366,971-6
Murakami, M. T. , Melo,C. C., Angulo,Y. , Lomonte,B. Arni, R. K. (2006).Acta Cryst.F62, 423-426
Yamazaki, Y., Matsunaga, Y., Nakano, Y. & Morita, T. (2005). J. Biol. Chem. 280, 29989–29992.
Created with the participation of Elie Peillon