Clp protease
From Proteopedia
| Line 1: | Line 1: | ||
[[Image:1tyf.png|left|200px]] | [[Image:1tyf.png|left|200px]] | ||
| - | <!-- | ||
| - | The line below this paragraph, containing "STRUCTURE_1tyf", creates the "Structure Box" on the page. | ||
| - | You may change the PDB parameter (which sets the PDB file loaded into the applet) | ||
| - | or the SCENE parameter (which sets the initial scene displayed when the page is loaded), | ||
| - | or leave the SCENE parameter empty for the default display. | ||
| - | --> | ||
{{STRUCTURE_1tyf| PDB=1tyf | SCENE= }} | {{STRUCTURE_1tyf| PDB=1tyf | SCENE= }} | ||
| Line 13: | Line 7: | ||
{{ABSTRACT_PUBMED_9390554}} | {{ABSTRACT_PUBMED_9390554}} | ||
| - | |||
| - | |||
| - | {{Link Toggle FancyCartoonHighQualityView}}. | ||
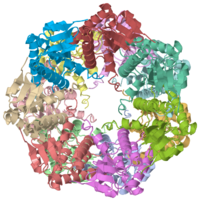
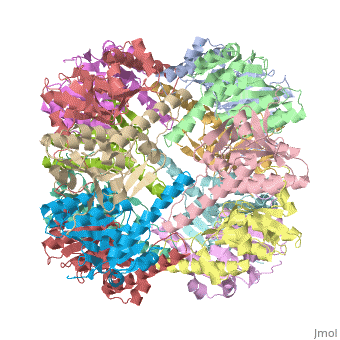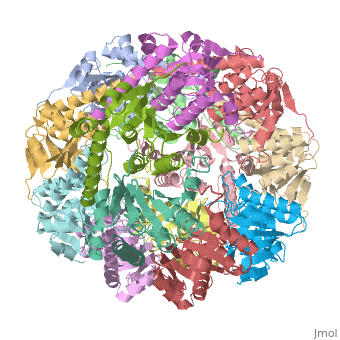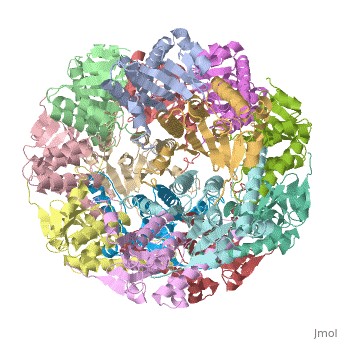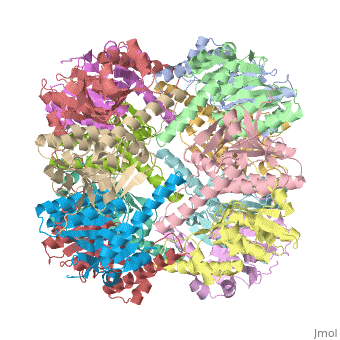
Two pores are found on each end of the peptidase <scene name='1tyf/1tyf_spacefill/2'>which are obvious when all proteins atoms shown as spheres with Van der Waals radii</scene>. | Two pores are found on each end of the peptidase <scene name='1tyf/1tyf_spacefill/2'>which are obvious when all proteins atoms shown as spheres with Van der Waals radii</scene>. | ||
Current revision
| |||||||||
| 1tyf, resolution 2.30Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Activity: | Endopeptidase Clp, with EC number 3.4.21.92 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
THE STRUCTURE OF CLPP AT 2.3 ANGSTROM RESOLUTION SUGGESTS A MODEL FOR ATP-DEPENDENT PROTEOLYSIS
We have determined the crystal structure of the proteolytic component of the caseinolytic Clp protease (ClpP) from E. coli at 2.3 A resolution using an ab initio phasing procedure that exploits the internal 14-fold symmetry of the oligomer. The structure of a ClpP monomer has a distinct fold that defines a fifth structural family of serine proteases but a conserved catalytic apparatus. The active protease resembles a hollow, solid-walled cylinder composed of two 7-fold symmetric rings stacked back-to-back. Its 14 proteolytic active sites are located within a central, roughly spherical chamber approximately 51 A in diameter. Access to the proteolytic chamber is controlled by two axial pores, each having a minimum diameter of approximately 10 A. From the structural features of ClpP, we suggest a model for its action in degrading proteins.
The structure of ClpP at 2.3 A resolution suggests a model for ATP-dependent proteolysis., Wang J, Hartling JA, Flanagan JM, Cell. 1997 Nov 14;91(4):447-56. PMID:9390554
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
Two pores are found on each end of the peptidase . In fact, one can even look view completely through the large lumen of the tetradecamer and out the other side of the structure
Reference
- Wang J, Hartling JA, Flanagan JM. The structure of ClpP at 2.3 A resolution suggests a model for ATP-dependent proteolysis. Cell. 1997 Nov 14;91(4):447-56. PMID:9390554
Created with the participation of Wayne Decatur.