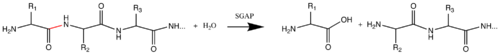Streptomyces griseus Aminopeptidase (SGAP)
From Proteopedia
(Difference between revisions)
| (5 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
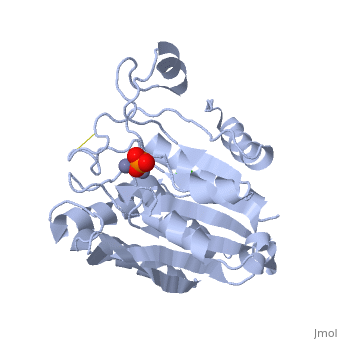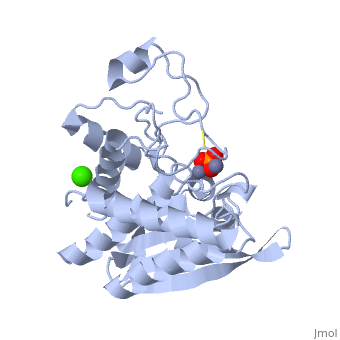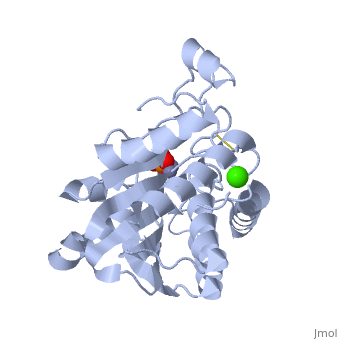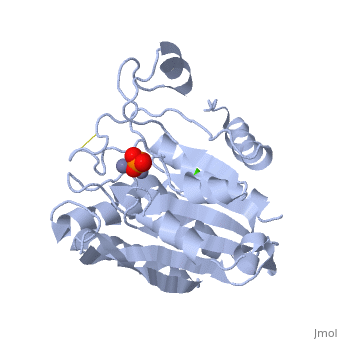
| - | + | <StructureSection load='1xjo' size='400' side='right' scene= caption='SGAP complex with oxymethionine, phosphate, Ca+2 (green) and Zn+2 (grey) (PDB code [[1xjo]])'> | |
| - | ''' | + | |
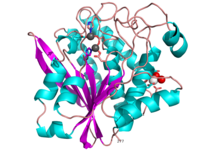
| + | [[Image:1xjo_cartoon.png |left| 200px | thumb | S griseus aminopeptidase, showing overall fold. Zinc ions are dark grey, calcium ion is white.]] | ||
== ''Streptomyces griseus'' Aminopeptidase (SGAP) == | == ''Streptomyces griseus'' Aminopeptidase (SGAP) == | ||
| Line 7: | Line 7: | ||
''S. griseus'' [[Aminopeptidase]] (SGAP; E.C. 3.4.11.-) cleaves the N-terminal amino acid from a peptide or protein, and is specific for larger hydrophobic acids, especially leucine. No cleavage occurs if the next residue is proline. | ''S. griseus'' [[Aminopeptidase]] (SGAP; E.C. 3.4.11.-) cleaves the N-terminal amino acid from a peptide or protein, and is specific for larger hydrophobic acids, especially leucine. No cleavage occurs if the next residue is proline. | ||
| - | [[Image:aminopeptidase_rxn2.png | left | thumb| | + | [[Image:aminopeptidase_rxn2.png | left | thumb| 500px | Reaction catalyzed by SGAP; scissile bond is shown in red.]] |
<br style="clear:both;"/> | <br style="clear:both;"/> | ||
==Structural features== | ==Structural features== | ||
| - | |||
| - | <!-- | ||
| - | The line below this paragraph, containing "STRUCTURE_ 1xjo", creates the "Structure Box" on the page. | ||
| - | You may change the PDB parameter (which sets the PDB file loaded into the applet) | ||
| - | or the SCENE parameter (which sets the initial scene displayed when the page is loaded), | ||
| - | or leave the SCENE parameter empty for the default display. | ||
| - | --> | ||
| - | {{STRUCTURE_ 1xjo | PDB= 1xjo | SCENE= }} | ||
SGAP belongs to the bacterial dinuclear zinc exopeptidase family and the Zn-dependent exopeptidase superfamily. It has a phosphorylase/hydrolase-like α/β fold, consisting of a 3-layer <scene name='1xjo/Sandwich/2'>αβα sandwich </scene>. | SGAP belongs to the bacterial dinuclear zinc exopeptidase family and the Zn-dependent exopeptidase superfamily. It has a phosphorylase/hydrolase-like α/β fold, consisting of a 3-layer <scene name='1xjo/Sandwich/2'>αβα sandwich </scene>. | ||
| Line 31: | Line 23: | ||
SGAP is one of the many proteinases present in the extracellular fluid of cultures of ''Streptomyces griseus'', and can be isolated from [http://en.wikipedia.org/wiki/Pronase Pronase], the commercial preparation of the extracellular fluid from this organism. SGAP is a monomeric, 30KDa, heat stable enzyme requiring two Zn<sup>2+</sup> ions for activity, and is activated by Ca<sup>2+</sup>. | SGAP is one of the many proteinases present in the extracellular fluid of cultures of ''Streptomyces griseus'', and can be isolated from [http://en.wikipedia.org/wiki/Pronase Pronase], the commercial preparation of the extracellular fluid from this organism. SGAP is a monomeric, 30KDa, heat stable enzyme requiring two Zn<sup>2+</sup> ions for activity, and is activated by Ca<sup>2+</sup>. | ||
| + | |||
| + | == Historical Context == | ||
| + | |||
| + | The proteolytic activity contained in the extracellular fluid of cultures of ''Streptomyces griseus'' was first identified by Nomoto and Narahashi (1959a), who obtained a highly purified preparation of this activity from the K-1 strain of this bactreria. A large scale version of their procedure was used to prepare commercial quantities of this preparation (Pronase). Various physical criteria showed that Pronase was homogeneous (Nomoto and Narahashi, 1959b), yet displayed both exopeptidase and endopeptidase activity, with a wide range of side chain specificities (Nomoto and Narahashi, 1959b, 1959c; Nomoto ''et al''., 1960a, 1960b, 1960c). The supposed homogeneity of Pronase was controversial, with other investigators using various chromatographic methods to isolate more fractions with proteolytic activity (Hiramatsu and Ouchi, 1963; Nomoto et al., 1964). Subsequently Narahashi and Yanagita (1967) identified several distinct proteolytic activities including one which had aminopeptidase activity, and was activated by Ca<sup>2+</sup> and Co<sup>2+</sup>. In contrast to other proteinase activities in the mixture, this aminopeptidase activity displayed considerable heat stablity (up to 80℃) and was unaffected by 9M urea. The activity, was, however, very sensitive to metal chelating agents. | ||
| + | |||
| + | While attempting to isolate the protein responsible for the trypsin activity in Pronase, Vosbeck ''et al.'' (1973) isolated two fractions with aminopeptidase activity. Although the two fractions differed slighty in their molecular weights (23K and 25K), they appeared to have the same enzymatic properties. | ||
| + | |||
| + | Interest in SGAP was renewed when aminopeptidases were recognized as useful tools in assays of metalloendopeptidase activity. The assay was based on a two stage reaction, with the endopeptidase cleaved an N-blocked peptide to release smaller peptide that was rapidly degraded by an aminopeptidase, generating a chromophore (Orlowski and Wilk, 1981; Mumford ''et al''., 1981). SGAP was considered an ideal tool for this purpose, given its stability, small size, and availability (Indig ''et al''., 1990). | ||
| + | |||
== Structures Available == | == Structures Available == | ||
| Line 45: | Line 46: | ||
* [[1tkj]] - with D-methionine to 1.15Å | * [[1tkj]] - with D-methionine to 1.15Å | ||
* [[1xbu]] - with ''p''-iodo-D-phenylalanine to 1.20Å | * [[1xbu]] - with ''p''-iodo-D-phenylalanine to 1.20Å | ||
| - | + | </StructureSection> | |
| - | == | + | ==See also== |
| - | + | [[Aminopeptidase]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
== References == | == References == | ||
<small> | <small> | ||
| - | + | * Greenblatt, H.M., Almog, O., Maras, B., Spungin-Bialik, A., Barra, D., Blumberg, S., Shoham, G., (1997) "Streptomyces griseus aminopeptidase: x-ray crystallographic structure at 1.75 a resolution", <i>J. Mol. Biol.</i> <b>265</b> (620). [http://www.ncbi.nlm.nih.gov/sites/entrez?db=pubmed&uid=9048953&cmd=showdetailview&indexed=google PMID: 9048953] | |
* Hiramatsu, A., & Ouchi, T. (1963). On the proteolytic enzymes from the commercial protease preparation of ''Streptomyces griseus'' (Pronase P). ''J. Biochem''. '''54''', 462-464. | * Hiramatsu, A., & Ouchi, T. (1963). On the proteolytic enzymes from the commercial protease preparation of ''Streptomyces griseus'' (Pronase P). ''J. Biochem''. '''54''', 462-464. | ||
* Indig, F.E., Benayahu, D., Fried, A., Wientroub, S., Blumberg, S. (1990). Neutral endopeptidase (EC 3.4.24.11) is highly expressed on osteoblastic cells and other marrow stromal cell types. ''Biochem. Biophys. Res. Commun''. '''172''', 620-626. [http://www.ncbi.nlm.nih.gov/pubmed/2241957?ordinalpos=1&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum PMID: 2241957] | * Indig, F.E., Benayahu, D., Fried, A., Wientroub, S., Blumberg, S. (1990). Neutral endopeptidase (EC 3.4.24.11) is highly expressed on osteoblastic cells and other marrow stromal cell types. ''Biochem. Biophys. Res. Commun''. '''172''', 620-626. [http://www.ncbi.nlm.nih.gov/pubmed/2241957?ordinalpos=1&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum PMID: 2241957] | ||
| Line 70: | Line 65: | ||
* Orlowski, M., & Wilk, S. (1981). Purification and specificity of a membrane-bound metalloendopeptidase from bovine pituitaries. ''Biochemistry'' '''20''', 4942-4950. [http://www.ncbi.nlm.nih.gov/pubmed/7028098?ordinalpos=12&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum PMID: 7028098] | * Orlowski, M., & Wilk, S. (1981). Purification and specificity of a membrane-bound metalloendopeptidase from bovine pituitaries. ''Biochemistry'' '''20''', 4942-4950. [http://www.ncbi.nlm.nih.gov/pubmed/7028098?ordinalpos=12&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum PMID: 7028098] | ||
* Vosbeck, K. D., Chow, K. F., & Awad, W. M. Jr (1973). The proteolytic enzymes of the K-1 strain of ''Streptomyces griseus'' obtained from a commercial preparation (Pronase). Purification and characterization of the aminopeptidases. '' J. Biol. Chem''. '''248''', 6029-6034. [http://www.ncbi.nlm.nih.gov/pubmed/4199257?ordinalpos=1&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum PMID: 4199257] | * Vosbeck, K. D., Chow, K. F., & Awad, W. M. Jr (1973). The proteolytic enzymes of the K-1 strain of ''Streptomyces griseus'' obtained from a commercial preparation (Pronase). Purification and characterization of the aminopeptidases. '' J. Biol. Chem''. '''248''', 6029-6034. [http://www.ncbi.nlm.nih.gov/pubmed/4199257?ordinalpos=1&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum PMID: 4199257] | ||
| - | |||
| - | --[[User:Harry|Harry]] 15:20, 17 March 2008 (IST) | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
See also
References
- Greenblatt, H.M., Almog, O., Maras, B., Spungin-Bialik, A., Barra, D., Blumberg, S., Shoham, G., (1997) "Streptomyces griseus aminopeptidase: x-ray crystallographic structure at 1.75 a resolution", J. Mol. Biol. 265 (620). PMID: 9048953
- Hiramatsu, A., & Ouchi, T. (1963). On the proteolytic enzymes from the commercial protease preparation of Streptomyces griseus (Pronase P). J. Biochem. 54, 462-464.
- Indig, F.E., Benayahu, D., Fried, A., Wientroub, S., Blumberg, S. (1990). Neutral endopeptidase (EC 3.4.24.11) is highly expressed on osteoblastic cells and other marrow stromal cell types. Biochem. Biophys. Res. Commun. 172, 620-626. PMID: 2241957
- Mumford, R.A., Pierzchala, P.A., Strauss, A.W., Zimmerman, M. (1981). Purification of a membrane-bound metalloendopeptidase from porcine kidney that degrades peptide hormones. Proc. Natl Acad. Sci. USA 78, 6623-6627. PMID: 7031658
- Narahashi, Y., & Yanagita, M. (1967). Studies on proteolytic enzymes (Pronase) of Streptomyces griseus K-1. I. Nature and properties of the proteolytic enzyme system. J. Biochem. (Tokyo) 62, 633-641. PMID: 4968616
- Nomoto, M., & Narahashi, Y. (1959a). A proteolytic enzyme of Streptomyces griseus: I. Purification of a protease of Streptomyces grisues. J. Biochem. 46, 653-667.
- Nomoto, M., & Narahashi, Y. (1959b). A proteolytic enzyme of Streptomyces griseus: III. Homogeneity of the purified enzyme preparation. J. Biochem. 46, 1481-1487.
- Nomoto, M., & Narahashi, Y. (1959c). A proteolytic enzyme of Streptomyces griseus: IV. General properties of Streptomyces grisues protease. J. Biochem. 46, 1645-1651.
- Nomoto, M., & Narahashi, Y., Murakami, M. (1960a). A proteolytic enzyme of Streptomyces griseus: V. Protective effect of calcium ion on the stability of protease. J. Biochem. 48, 453-463.
- Nomoto, M., & Narahashi, Y., Murakami, M. (1960b). A proteolytic enzyme of Streptomyces griseus: VI. Hydrolysis of protein by Streptomyces griseus protease. J. Biochem. 48, 593-602.
- Nomoto, M., & Narahashi, Y., Murakami, M. (1960c). A proteolytic enzyme of Streptomyces griseus: VII. Substrate specificity of Streptomyces griseus protease. J. Biochem. 48, 906-918.
- Nomoto, M., Narahashi, Y., Ouchi, T., & Hiramatsu, A. (1964). Abstract, 6th Inern. Congr. Biochem., N.Y., 4, 123.
- Orlowski, M., & Wilk, S. (1981). Purification and specificity of a membrane-bound metalloendopeptidase from bovine pituitaries. Biochemistry 20, 4942-4950. PMID: 7028098
- Vosbeck, K. D., Chow, K. F., & Awad, W. M. Jr (1973). The proteolytic enzymes of the K-1 strain of Streptomyces griseus obtained from a commercial preparation (Pronase). Purification and characterization of the aminopeptidases. J. Biol. Chem. 248, 6029-6034. PMID: 4199257
Proteopedia Page Contributors and Editors (what is this?)
Harry Greenblatt, Alexander Berchansky, Michal Harel, Eric Martz