Sandbox SouthUniversity1
From Proteopedia
(Difference between revisions)
| (One intermediate revision not shown.) | |||
| Line 9: | Line 9: | ||
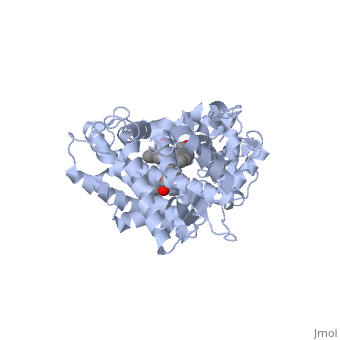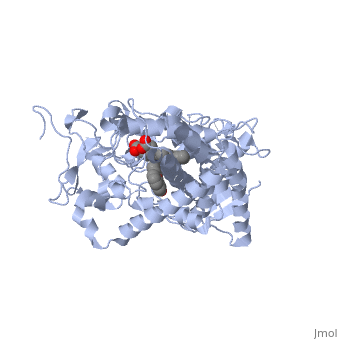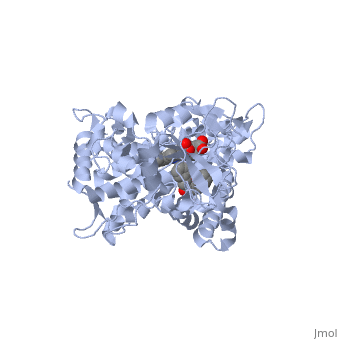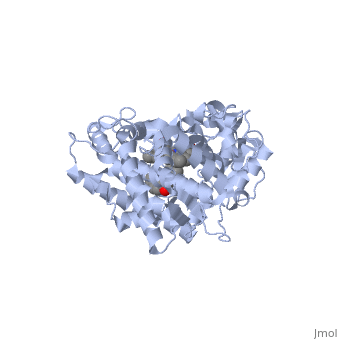
First, lets look at the heme. Click the following link to hide the rest of the protein and <scene name='Sandbox_SouthUniversity1/Heme_only/1'>highlight the heme ring system.</scene> Carbon atoms are shown in grey, Nitrogen atoms are blue, Oxygen is red, and the central iron atom is orange. The iron atom is a vital center for oxidation of substrates (drugs or other xenobiotics). It is shown <scene name='Sandbox_SouthUniversity1/Heme_sticks_fe_ball/1'>here,</scene> with the protein strands displayed partially transparent to highlight the heme ring system and iron atom. | First, lets look at the heme. Click the following link to hide the rest of the protein and <scene name='Sandbox_SouthUniversity1/Heme_only/1'>highlight the heme ring system.</scene> Carbon atoms are shown in grey, Nitrogen atoms are blue, Oxygen is red, and the central iron atom is orange. The iron atom is a vital center for oxidation of substrates (drugs or other xenobiotics). It is shown <scene name='Sandbox_SouthUniversity1/Heme_sticks_fe_ball/1'>here,</scene> with the protein strands displayed partially transparent to highlight the heme ring system and iron atom. | ||
| - | Now look at the substrate molecule being oxidized and view its orientation relative to the heme group by clicking <scene name='Sandbox_SouthUniversity1/Heme_and_flavone/3'>this link</scene> Resize and rotate the molecules until you can see how the two molecules are oriented in relationship to each other. The flavone is metabolized (oxidized) by introduction of a hydroxy group onto the phenyl ring that is attached to the tricyclic ring system. | + | Now look at the substrate molecule being oxidized and view its orientation relative to the heme group by clicking <scene name='Sandbox_SouthUniversity1/Heme_and_flavone/3'>this link</scene> Resize and rotate the molecules until you can see how the two molecules are oriented in relationship to each other. The flavone is metabolized (oxidized) by introduction of a hydroxy group onto the phenyl ring that is attached to the tricyclic ring system. Note particularly the distance and orientation of the heme iron and the phenyl group. |
| Line 15: | Line 15: | ||
| - | The substrate (flavone) shown here is different from many other substrates of this CYP. Besides being a substrate of CYP1A2, it is also an inhibitor. It inhibits the metabolism of other CYP1A2 substrates because it binds very tightly to this enzyme. Thus, while the CYP is bound, it is unavailable to metabolize other substrates (e.g. drugs). Therefor, if a drug were | + | The '''substrate''' (flavone) shown here is different from many other substrates of this CYP. Besides being a substrate of CYP1A2, it is also an '''inhibitor'''. It inhibits the metabolism of other CYP1A2 substrates because it binds very tightly to this enzyme. Thus, while the CYP is bound, it is unavailable to metabolize other substrates (e.g. drugs). Therefor, if a drug were coadministered with this compound, it might not be broken down to the extent expected, and could build up to toxic levels. |
| - | The reason that this flavone is bound so tightly to the enzyme is that it's shape and | + | The reason that this flavone is bound so tightly to the enzyme is that it's shape and electronic charge are complementary to the binding pocket. This is examined <scene name='Sandbox_SouthUniversity1/2hi4_blue_transparent_ribbons/1'>next.</scene> |
| + | |||
| + | First examine the shape of the VDW area <scene name='Sandbox_SouthUniversity1/2hi4_bl_transp_rib_flav_vdw/1'>around the flavone.</scene> The reddish areas show places where there are particularly close contacts with the binding pocket. These contacts can be caused by ionic, hydrophobic, or hydrogen bonding. Now we will examine what exactly is responsible for the tight binding. | ||
| + | |||
| + | Lets remove the rest of the protein, so we can better see these interactions. | ||
| + | |||
| + | First examine the shape of the <scene name='Sandbox_SouthUniversity1/2hi4_bl_transp_ribbon_flav_spc/1'>flavone </scene>bound to the protein. | ||
| + | Next, the Van derWaals surface of the cavity is <scene name='Sandbox_SouthUniversity1/displayed./1'>TextToBeDisplayed</scene> | ||
Different members of the CYP450 enzymes preferentially metabolize different xenobiotics. What features of a drug do you think might cause it to be metabolized by one CYP versus another? | Different members of the CYP450 enzymes preferentially metabolize different xenobiotics. What features of a drug do you think might cause it to be metabolized by one CYP versus another? | ||
Current revision
==Selectivity of Drug Metabolism by Cytochrome P450 Enzymes==
| |||||||||||
Draft: <illustration of residues in active site> Draft animation Draft animation
draft: <insert quiz here on electrostatic and steric factors>
draft: <insert animations here contrasting the shape of the active site of CYP3A4 and e.g. CYP2E1>
draft: <insert examples of substrates for 2 different CYPs, overlay them on top of each other illustrate the molecular commonalities>
draft <illustrate the size of the binding pockets and their electronic character>
draft test of scene names:
<Draft Quiz>
References The structure of CYP1A2 shown above is found in the Research Collaboration of Structural Bioinformatics Protein Database as entry 2HI4.