We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
1mkf
From Proteopedia
(Difference between revisions)
| (8 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | [[Image:1mkf.png|left|200px]] | ||
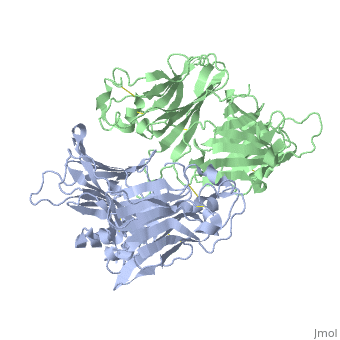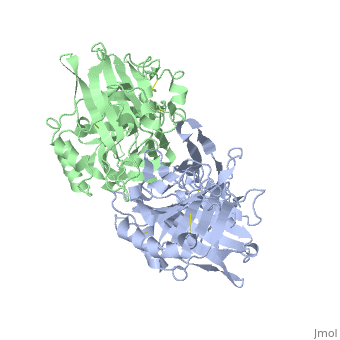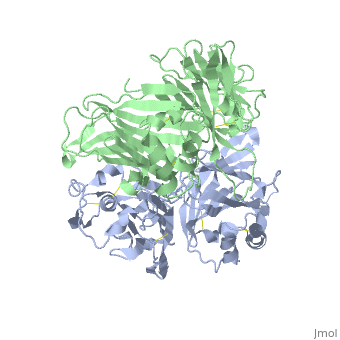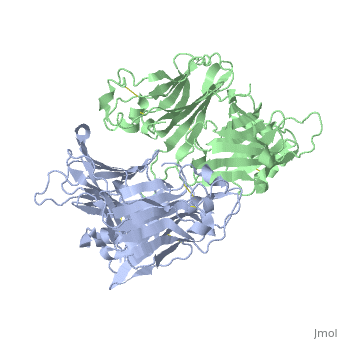
| - | < | + | ==VIRAL CHEMOKINE BINDING PROTEIN M3 FROM MURINE GAMMAHERPESVIRUS 68== |
| - | + | <StructureSection load='1mkf' size='340' side='right'caption='[[1mkf]], [[Resolution|resolution]] 2.10Å' scene=''> | |
| - | + | == Structural highlights == | |
| - | + | <table><tr><td colspan='2'>[[1mkf]] is a 2 chain structure with sequence from [https://en.wikipedia.org/wiki/Murid_gammaherpesvirus_4 Murid gammaherpesvirus 4]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1MKF OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1MKF FirstGlance]. <br> | |
| - | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.1Å</td></tr> | |
| - | -- | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1mkf FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1mkf OCA], [https://pdbe.org/1mkf PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1mkf RCSB], [https://www.ebi.ac.uk/pdbsum/1mkf PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1mkf ProSAT], [https://www.topsan.org/Proteins/MCSG/1mkf TOPSAN]</span></td></tr> |
| - | + | </table> | |
| + | == Function == | ||
| + | [https://www.uniprot.org/uniprot/O41925_MHV68 O41925_MHV68] | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
| + | The M3 protein encoded by murine gamma herpesvirus68 (gamma HV68) functions as an immune system saboteur by the engagement of chemoattractant cytokines, thereby altering host antiviral inflammatory responses. Here we report the crystal structures of M3 both alone and in complex with the CC chemokine MCP-1. M3 is a two-domain beta sandwich protein with a unique sequence and topology, forming a tightly packed anti-parallel dimer. The stoichiometry of the MCP-1:M3 complex is 2:2, with two monomeric chemokines embedded at distal ends of the preassociated M3 dimer. Conformational flexibility and electrostatic complementation are both used by M3 to achieve high-affinity and broad-spectrum chemokine engagement. M3 also employs structural mimicry to promiscuously sequester chemokines, engaging conservative structural elements associated with both chemokine homodimerization and binding to G protein-coupled receptors. | ||
| - | + | Structural basis of chemokine sequestration by a herpesvirus decoy receptor.,Alexander JM, Nelson CA, van Berkel V, Lau EK, Studts JM, Brett TJ, Speck SH, Handel TM, Virgin HW, Fremont DH Cell. 2002 Nov 1;111(3):343-56. PMID:12419245<ref>PMID:12419245</ref> | |
| - | + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |
| - | + | </div> | |
| - | + | <div class="pdbe-citations 1mkf" style="background-color:#fffaf0;"></div> | |
| - | + | == References == | |
| - | + | <references/> | |
| - | + | __TOC__ | |
| - | + | </StructureSection> | |
| - | == | + | [[Category: Large Structures]] |
| - | + | [[Category: Murid gammaherpesvirus 4]] | |
| - | + | [[Category: Alexander JM]] | |
| - | + | [[Category: Fremont DH]] | |
| - | + | ||
| - | + | ||
| - | == | + | |
| - | < | + | |
| - | [[Category: | + | |
| - | [[Category: | + | |
| - | [[Category: | + | |
| - | [[Category: | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
VIRAL CHEMOKINE BINDING PROTEIN M3 FROM MURINE GAMMAHERPESVIRUS 68
| |||||||||||