This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
CED-4 Apoptosome
From Proteopedia
(Difference between revisions)
(New page: 200px <!-- The line below this paragraph, containing "STRUCTURE_3lqq", creates the "Structure Box" on the page. You may change the PDB parameter (which sets the PD...) |
|||
| (4 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| + | <StructureSection load="3lqq" size="350" color="" frame="true" spin="on" Scene='3lqq/Default/1' side="right" caption="Nematode CED-4 apoptosome complex with ATP and Mg+2 ion [[3lqq]]" > | ||
[[Image:3lqq.jpg|left|200px]] | [[Image:3lqq.jpg|left|200px]] | ||
| - | + | ===Structure of the CED-4 Apoptosome ([[3lqq]])=== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ===Structure of the CED-4 Apoptosome=== | + | |
{{ABSTRACT_PUBMED_20434985}} | {{ABSTRACT_PUBMED_20434985}} | ||
<!-- | <!-- | ||
| Line 24: | Line 15: | ||
==About this Structure== | ==About this Structure== | ||
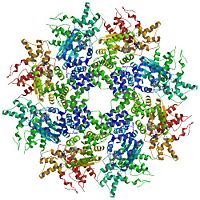
| - | + | [[3lqq]] is a 2 chains structure with sequences from [http://en.wikipedia.org/wiki/Caenorhabditis_elegans Caenorhabditis elegans]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3LQQ OCA]. | |
<scene name='3lqq/Default/1'>The crystallographic asymmetric unit of the solved structure is an asymmetric dimer</scene> of CED-4. <scene name='3lqq/Morph/2'>Morph showing how each monomer in the asymmetric dimer relate to each other</scene>. Be sure to spin it around because <scene name='3lqq/Morphside/3'>from the side it is easier to see the dramatic difference</scene>.<br> {{Button Toggle AnimationOnPause}}<!--When Proteopedia updates to a version of Jmol that supports the Compare command, I should upload a new pdb file with chain as a model and chain b as a model in a single pdb file and then replace this scene with a scripted compare that just compares model 2 (chain b) to model 1 (chain a).--> | <scene name='3lqq/Default/1'>The crystallographic asymmetric unit of the solved structure is an asymmetric dimer</scene> of CED-4. <scene name='3lqq/Morph/2'>Morph showing how each monomer in the asymmetric dimer relate to each other</scene>. Be sure to spin it around because <scene name='3lqq/Morphside/3'>from the side it is easier to see the dramatic difference</scene>.<br> {{Button Toggle AnimationOnPause}}<!--When Proteopedia updates to a version of Jmol that supports the Compare command, I should upload a new pdb file with chain as a model and chain b as a model in a single pdb file and then replace this scene with a scripted compare that just compares model 2 (chain b) to model 1 (chain a).--> | ||
<scene name='3lqq/Biomolecule1/4'>The biological molecule is an octamer (tetramer of the asymmetric dimer)</scene> that forms a shape similar to a funnel.<br> | <scene name='3lqq/Biomolecule1/4'>The biological molecule is an octamer (tetramer of the asymmetric dimer)</scene> that forms a shape similar to a funnel.<br> | ||
| - | + | ||
==External Links== | ==External Links== | ||
* [http://www.nature.com/nchina/2010/100707/full/nchina.2010.77.html '''Structural biology: The funnel of death''' by Felix Cheung] at Nature China. | * [http://www.nature.com/nchina/2010/100707/full/nchina.2010.77.html '''Structural biology: The funnel of death''' by Felix Cheung] at Nature China. | ||
| Line 45: | Line 36: | ||
[[Category: Mitochondrion]] | [[Category: Mitochondrion]] | ||
[[Category: Nucleotide-binding]] | [[Category: Nucleotide-binding]] | ||
| + | <br /> | ||
| + | Created with the participation of [[User:Wayne Decatur|Wayne Decatur]], [[User:Eran Hodis|Eran Hodis]]. | ||
| + | ==See also== | ||
| + | [[Cell death protein]] | ||
Current revision
| |||||||||||
Categories: Caenorhabditis elegans | Liu, Q. | Pang, Y. | Qi, S. | Shi, Y. | Yan, N. | Alternative splicing | Apoptosis | Apoptosome | Atp-binding | Ced4 | Mitochondrion | Nucleotide-binding