We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
3kyc
From Proteopedia
(Difference between revisions)
m (Protected "3kyc" [edit=sysop:move=sysop]) |
|||
| (10 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | [[Image:3kyc.png|left|200px]] | ||
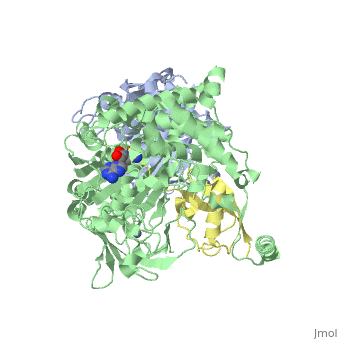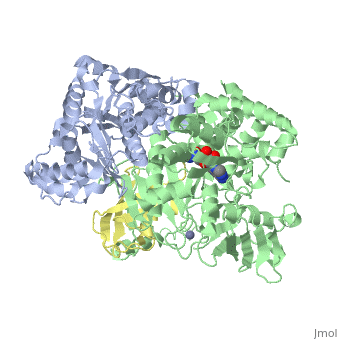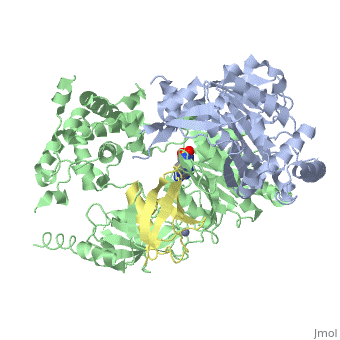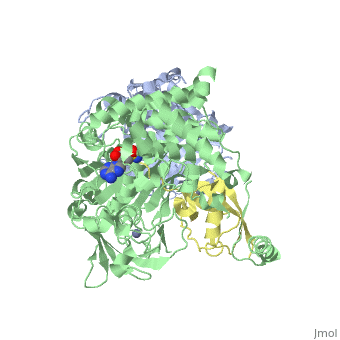
| - | < | + | ==Human SUMO E1 complex with a SUMO1-AMP mimic== |
| - | + | <StructureSection load='3kyc' size='340' side='right'caption='[[3kyc]], [[Resolution|resolution]] 2.45Å' scene=''> | |
| - | You may | + | == Structural highlights == |
| - | + | <table><tr><td colspan='2'>[[3kyc]] is a 3 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3KYC OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=3KYC FirstGlance]. <br> | |
| - | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.45Å</td></tr> | |
| - | + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=JZU:5-DEOXY-5-(SULFAMOYLAMINO)ADENOSINE'>JZU</scene>, <scene name='pdbligand=ZN:ZINC+ION'>ZN</scene></td></tr> | |
| - | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=3kyc FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3kyc OCA], [https://pdbe.org/3kyc PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=3kyc RCSB], [https://www.ebi.ac.uk/pdbsum/3kyc PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=3kyc ProSAT]</span></td></tr> | |
| + | </table> | ||
| + | == Function == | ||
| + | [https://www.uniprot.org/uniprot/SAE2_HUMAN SAE2_HUMAN] The heterodimer acts as a E1 ligase for SUMO1, SUMO2, SUMO3, and probably SUMO4. It mediates ATP-dependent activation of SUMO proteins followed by formation of a thioester bond between a SUMO protein and a conserved active site cysteine residue on UBA2/SAE2.<ref>PMID:11481243</ref> <ref>PMID:11451954</ref> <ref>PMID:19443651</ref> <ref>PMID:15660128</ref> <ref>PMID:17643372</ref> <ref>PMID:20164921</ref> | ||
| + | == Evolutionary Conservation == | ||
| + | [[Image:Consurf_key_small.gif|200px|right]] | ||
| + | Check<jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/ky/3kyc_consurf.spt"</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview03.spt</scriptWhenUnchecked> | ||
| + | <text>to colour the structure by Evolutionary Conservation</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=3kyc ConSurf]. | ||
| + | <div style="clear:both"></div> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
| + | E1 enzymes activate ubiquitin (Ub) and ubiquitin-like (Ubl) proteins in two steps by carboxy-terminal adenylation and thioester bond formation to a conserved catalytic cysteine in the E1 Cys domain. The structural basis for these intermediates remains unknown. Here we report crystal structures for human SUMO E1 in complex with SUMO adenylate and tetrahedral intermediate analogues at 2.45 and 2.6 A, respectively. These structures show that side chain contacts to ATP.Mg are released after adenylation to facilitate a 130 degree rotation of the Cys domain during thioester bond formation that is accompanied by remodelling of key structural elements including the helix that contains the E1 catalytic cysteine, the crossover and re-entry loops, and refolding of two helices that are required for adenylation. These changes displace side chains required for adenylation with side chains required for thioester bond formation. Mutational and biochemical analyses indicate these mechanisms are conserved in other E1s. | ||
| - | + | Active site remodelling accompanies thioester bond formation in the SUMO E1.,Olsen SK, Capili AD, Lu X, Tan DS, Lima CD Nature. 2010 Feb 18;463(7283):906-12. PMID:20164921<ref>PMID:20164921</ref> | |
| - | + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |
| - | + | </div> | |
| - | + | <div class="pdbe-citations 3kyc" style="background-color:#fffaf0;"></div> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | |
| - | + | ||
==See Also== | ==See Also== | ||
*[[Human SUMO E1 complex|Human SUMO E1 complex]] | *[[Human SUMO E1 complex|Human SUMO E1 complex]] | ||
| - | *[[Human SUMO E1 complex with a SUMO1-AMP mimic|Human SUMO E1 complex with a SUMO1-AMP mimic]] | ||
| - | *[[Human SUMO E1~SUMO1-AMP tetrahedral intermediate mimic|Human SUMO E1~SUMO1-AMP tetrahedral intermediate mimic]] | ||
*[[SUMO|SUMO]] | *[[SUMO|SUMO]] | ||
| - | + | *[[SUMO 3D Structures|SUMO 3D Structures]] | |
| - | == | + | *[[3D structures of Ubiquitin activating enzyme|3D structures of Ubiquitin activating enzyme]] |
| - | < | + | == References == |
| + | <references/> | ||
| + | __TOC__ | ||
| + | </StructureSection> | ||
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
| - | [[Category: | + | [[Category: Large Structures]] |
| - | [[Category: | + | [[Category: Lima CD]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
Human SUMO E1 complex with a SUMO1-AMP mimic
| |||||||||||