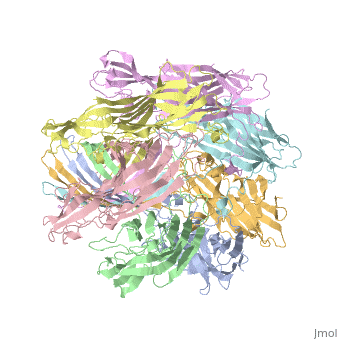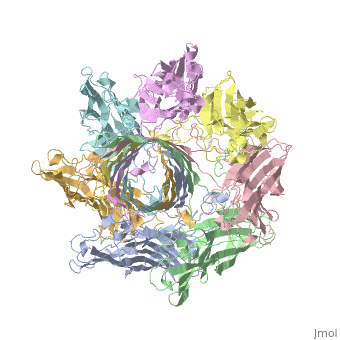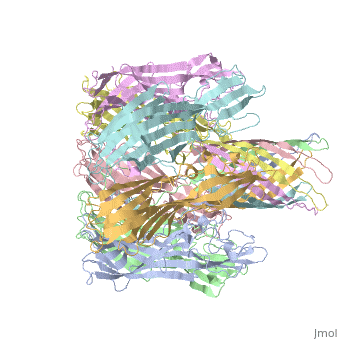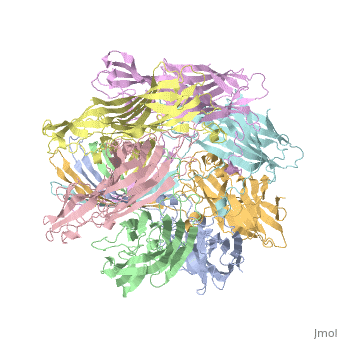We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Hemolysin
From Proteopedia
(Difference between revisions)
m (→α-hemolysin) |
m |
||
| (23 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='7ahl' size='350' side='right' caption='α-hemolysin heptamer (PDB code [[7ahl]]).' scene=''> | |
| + | == Function == | ||
| + | '''Hemolysin''' (HL) is exotoxin from bacteria which causes lysis of red blood cells<ref>PMID:20110774</ref>. | ||
| + | *'''alpha-hemolysin''' is a transmembrane pore-forming heptameric molecule<ref>PMID:8943190</ref>. See details for in [[Pore forming toxin, α-hemolysin]]. | ||
| + | *'''delta-hemolysin''' is a 26 amino acid peptide from the bacterium ''Staphylococcus'' exhibiting antimicrobial activity against'' Legionerlla'' <ref>PMID:19150639</ref>. | ||
| - | + | See details of hemolysin E in [[Molecular Playground/ClyA]]. | |
| - | + | For toxins in Proteopdia see [[Toxins]]. | |
| - | == | + | == Relevance == |
| + | HL acts as a virulence factor in the pathogenesis of invasive infections<ref>PMID:12564994</ref>. | ||
| - | + | ==3D Printed Physical Model of Hemolysin== | |
| - | + | Shown below is a 3D printed physical model of Hemolysin. The model is shown in alpha carbon backbone format with each chain colored uniquely. | |
| - | [[ | + | [[Image:hemolysin1_centerForBioMolecularModeling.jpg|550px]] |
| - | + | [[Image:hemolysin2_centerForBioMolecularModeling.jpg|550px]] | |
| - | + | ||
| - | + | ||
| - | === | + | ====The MSOE Center for BioMolecular Modeling==== |
| - | [[ | + | [[Image:CbmUniversityLogo.jpg | left | 150px]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | The [http://cbm.msoe.edu MSOE Center for BioMolecular Modeling] uses 3D printing technology to create physical models of protein and molecular structures, making the invisible molecular world more tangible and comprehensible. To view more protein structure models, visit our [http://cbm.msoe.edu/educationalmedia/modelgallery/ Model Gallery]. | |
| - | + | == 3D Structures of hemolysin == | |
| - | + | ||
| - | + | ||
| - | == | + | |
| - | + | ||
| - | + | ||
| - | + | [[Hemolysin 3D structures]] | |
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | == References == | ||
| + | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
| + | [[Category:3D printer files]] | ||
Current revision
| |||||||||||
References
- ↑ Mestre MB, Fader CM, Sola C, Colombo MI. Alpha-hemolysin is required for the activation of the autophagic pathway in Staphylococcus aureus-infected cells. Autophagy. 2010 Jan;6(1):110-25. PMID:20110774
- ↑ Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996 Dec 13;274(5294):1859-66. PMID:8943190
- ↑ Verdon J, Girardin N, Lacombe C, Berjeaud JM, Héchard Y. delta-hemolysin, an update on a membrane-interacting peptide. Peptides. 2009 Apr;30(4):817-23. PMID:19150639 doi:10.1016/j.peptides.2008.12.017
- ↑ Nizet V. Streptococcal beta-hemolysins: genetics and role in disease pathogenesis. Trends Microbiol. 2002 Dec;10(12):575-80. PMID:12564994
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Mark Hoelzer, Wayne Decatur, Alexander Berchansky, Marius Mihasan