Human beta two microglobulin
From Proteopedia
(Difference between revisions)
| (One intermediate revision not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='1dq8' size=' | + | <StructureSection load='1dq8' size='400' side='right' caption='Crystal Structure of the Human Beta-2 Microglobulin (PDB entry [[1lds]])' scene=''> |
===Beta two microglubulin in human class I major histocompatibility complex (MHCb2m)=== | ===Beta two microglubulin in human class I major histocompatibility complex (MHCb2m)=== | ||
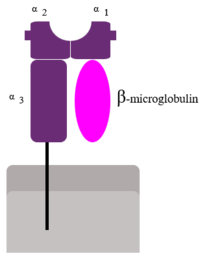
| - | [[Image:MHC Class 1.png|200px|left]]Human β2-Microglobulin is the non-covalently bound <scene name='Human_beta_two_microglobulin/Light_chain/2'>light chain</scene> | + | [[Image:MHC Class 1.png|200px|left]] |
| + | {{Clear}} | ||
| + | Human β2-Microglobulin is the non-covalently bound <scene name='Human_beta_two_microglobulin/Light_chain/2'>light chain</scene> | ||
of the human class I | of the human class I | ||
major histocompatibility complex (MHC-I). Its function is to ensure proper folding and cell-surface expression of MHC-1. | major histocompatibility complex (MHC-I). Its function is to ensure proper folding and cell-surface expression of MHC-1. | ||
| Line 17: | Line 19: | ||
and D. The most significant difference in the ctrystal structures of Mhb2m and MHCb2m involves residues in β strand D and the succeeding loop. When complexed with the MHC heavy chain, residues 50-56 of MHCb2m form two short β strands that separated by a two residue β bulge. These strands (depicted as D1 and D2 in Fig.1) each forms three main-chain-main-chain hydrogen bonds to the adjacent β strand E. The bulge in MHCb2m effectively twists the edge strand, which facilitate its binding to the surface of the heavy chain. However, this β bulge no longer exits in the crytal strucure of Mhb2m. The <scene name='Human_beta_two_microglobulin/D_strand/1'>conformation of D strand </scene>in Mhb2m provides an ideal assembly surface, making this edge-strand pair vulnerable to aggregation. The hydrogen-bonding potential of strand D is satisfied by the formation intermolecular interactionswith adjecent molecules, demonstrating the potential for this region to propagate assembly through edge-strand interactions. | and D. The most significant difference in the ctrystal structures of Mhb2m and MHCb2m involves residues in β strand D and the succeeding loop. When complexed with the MHC heavy chain, residues 50-56 of MHCb2m form two short β strands that separated by a two residue β bulge. These strands (depicted as D1 and D2 in Fig.1) each forms three main-chain-main-chain hydrogen bonds to the adjacent β strand E. The bulge in MHCb2m effectively twists the edge strand, which facilitate its binding to the surface of the heavy chain. However, this β bulge no longer exits in the crytal strucure of Mhb2m. The <scene name='Human_beta_two_microglobulin/D_strand/1'>conformation of D strand </scene>in Mhb2m provides an ideal assembly surface, making this edge-strand pair vulnerable to aggregation. The hydrogen-bonding potential of strand D is satisfied by the formation intermolecular interactionswith adjecent molecules, demonstrating the potential for this region to propagate assembly through edge-strand interactions. | ||
| - | In addition, the changes observed in strand D result in differenr orientations of the side chains of residues 50-54. As a result, | ||
| - | His-51 (which points inwards in the structure of MHCb2m) rotates by approximately 180, such that it now points away from the hydrophobic core of the protein. This would remove the second protevtive feature from the edge strand, facilitating further interaction in this region (Fig.2). | ||
| - | + | In addition, the changes observed in strand D result in differenr orientations of the side chains of residues 50-54. As a result, His-51 (which points inwards in the structure of MHCb2m) rotates by approximately 180, such that it now points away from the hydrophobic core of the protein. This would remove the second protevtive feature from the edge strand, facilitating further interaction in this region (Fig.2). | |
| + | |||
| + | |||
__NOTOC__ | __NOTOC__ | ||
=Amyloid Fibril formation of Mhb2m= | =Amyloid Fibril formation of Mhb2m= | ||
| Line 52: | Line 54: | ||
#.An additional common characteristic is that many variants capable of fibril formation at neutral pH have the effect of destabilising the Nterminal region of the protein, which is also highly unfolded in the structural ensembles formed at low pH. | #.An additional common characteristic is that many variants capable of fibril formation at neutral pH have the effect of destabilising the Nterminal region of the protein, which is also highly unfolded in the structural ensembles formed at low pH. | ||
#. The role of trans Pro32 in fibril formation at low pH is currently unknown, although 80% of the molecules would be expected to contain the trans conformation in the acid denatured state. Moreover, the observation that the rate of fibril formation of P32G is similar to that of wild-type b2m when studied at pH 2.5 is suggestive of a common trans amyloid precursor. | #. The role of trans Pro32 in fibril formation at low pH is currently unknown, although 80% of the molecules would be expected to contain the trans conformation in the acid denatured state. Moreover, the observation that the rate of fibril formation of P32G is similar to that of wild-type b2m when studied at pH 2.5 is suggestive of a common trans amyloid precursor. | ||
| - | + | </StructureSection> | |
==3D structures of beta-2 microglobulin== | ==3D structures of beta-2 microglobulin== | ||
Current revision
| |||||||||||
3D structures of beta-2 microglobulin
References
- Amyloid formation by globular proteins under native conditions. Nature chemical biology,5,15(2009) [1]
- Crystal structure of monomeric human β-2-microglobulin reveals clues to its amyloidogenic properties. PNAS,99,9771(2002) [2]
- A regulatable switch mediates self-association in an immunoglobulin fold. Nature structural & molecular biology,15,965(2008) [3]
- A native to amyloidogenic transition regulated by a beckbone trigger. Nature structural & molecular biology, 13,202(2006) [4]
- Glimpse of the molecular mechanism of β2-microglobulin fibril formation in vitro: Aggregation on a complex energy landscape. FEBS letters, 583,2623(2009) [5]