We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Papain
From Proteopedia
(Difference between revisions)
| (22 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | [[Image:papayas.jpg| | + | <StructureSection load='9pap' size='350' side='right' scene='Papain/Primary_scene/2' caption='Click on the links to the left to view different structural aspects of papain. |
| - | Papain. Meat tenderizer. Old time home remedy for insect, jellyfish, and stingray stings<ref>[http://www.ameriden.com/products/advanced-digestive-enzyme/] Ameridan International</ref>. Who would have thought that a sulfhydryl protease from the latex of the papaya fruit, ''Carica papaya'' and ''Vasconcellea cundinamarcensis'', would have such a practical application beyond Proteopedia? | + | PDB code [[9pap]]'> |
| + | [[Image:papayas.jpg|200px|left|thumb|Papaya<ref>[http://dailyfitnessmagz.com/2011/03/papayas-nutrition-facts/] Papaya's Nutrition Facts</ref>]] | ||
| + | |||
| + | == Introduction == | ||
| + | '''Papain'''. Meat tenderizer. Old time home remedy for insect, jellyfish, and stingray stings<ref>[http://www.ameriden.com/products/advanced-digestive-enzyme/] Ameridan International</ref>. Who would have thought that a sulfhydryl protease from the latex of the papaya fruit, ''Carica papaya'' and ''Vasconcellea cundinamarcensis'', would have such a practical application beyond Proteopedia? | ||
Papain is a 23.4 kDa, 212 residue cysteine protease, also known as '''papaya proteinase I''', from the peptidase C1 family ([[EC Number|E.C.]] [[Hydrolase|3.4.22.2]]).<ref>[http://www.uniprot.org/uniprot/P00784] Uniprot</ref><ref name="9PAP PDB">[http://www.pdb.org/pdb/explore/explore.do?structureId=9PAP] 9PAP PDB</ref> It is the natural product of the [http://en.wikipedia.org/wiki/Carica_papaya Papaya](''Carica papaya'')<ref name="Sigma Aldrich">[http://www.sigmaaldrich.com/life-science/metabolomics/enzyme-explorer/analytical-enzymes/papain.html] Sigma Aldrich</ref>, and may be extracted from the plant's latex, leaves and roots. Papain displays a broad range of functions, acting as an endopeptidase, exopeptidase, amidase, and esterase,<ref name="Worthington">[http://www.worthington-biochem.com/pap/default.html] Worthington</ref> with its optimal activity values for pH lying between 6.0 and 7.0, and its optimal temperature for activity is 65 °C. Its pI values are 8.75 and 9.55, and it is best visualized at a wavelength of 278 nm. <ref name="Sigma Aldrich" /> | Papain is a 23.4 kDa, 212 residue cysteine protease, also known as '''papaya proteinase I''', from the peptidase C1 family ([[EC Number|E.C.]] [[Hydrolase|3.4.22.2]]).<ref>[http://www.uniprot.org/uniprot/P00784] Uniprot</ref><ref name="9PAP PDB">[http://www.pdb.org/pdb/explore/explore.do?structureId=9PAP] 9PAP PDB</ref> It is the natural product of the [http://en.wikipedia.org/wiki/Carica_papaya Papaya](''Carica papaya'')<ref name="Sigma Aldrich">[http://www.sigmaaldrich.com/life-science/metabolomics/enzyme-explorer/analytical-enzymes/papain.html] Sigma Aldrich</ref>, and may be extracted from the plant's latex, leaves and roots. Papain displays a broad range of functions, acting as an endopeptidase, exopeptidase, amidase, and esterase,<ref name="Worthington">[http://www.worthington-biochem.com/pap/default.html] Worthington</ref> with its optimal activity values for pH lying between 6.0 and 7.0, and its optimal temperature for activity is 65 °C. Its pI values are 8.75 and 9.55, and it is best visualized at a wavelength of 278 nm. <ref name="Sigma Aldrich" /> | ||
| - | Papain's enzymatic use was first discovered in 1873 by G.C. Roy who published his results in the Calcutta Medical Journal in the article, "The Solvent Action of Papaya Juice on Nitrogenous Articles of Food." In 1879, Papain was named officially by Wurtz and Bouchut, who managed to partially purify the product from the sap of papaya. It wasn't until the mid-twentieth century that the complete purification and isolation of Papain was achieved. In 1968, Drenth et al. determined the structure of Papain by x-ray crystallography, making it the second enzyme whose structure was successfully determined by x-ray crystallography. Additionally, Papain was the first cysteine protease to have its structure identified.<ref name="Worthington" /> In 1984, Kamphuis et al. determined the geometry of the active site, and the three-dimensional structure was visualized to a 1.65 Angstrom solution.<ref name="Structure">PMID:6502713</ref> Today, studies continue on the stability of Papain, involving changes in environmental conditions as well as testing of inhibitors such as phenylmethanesulfonylfluoride (PMSF), TLCK, TPCK, aplh2-macroglobulin, heavy metals, AEBSF, antipain, cystatin, E-64, leupeptin, sulfhydryl binding agents, carbonyl reagents, and alkylating agents.<ref name="Worthington" /> | + | Papain's enzymatic use was first discovered in 1873 by G.C. Roy who published his results in the Calcutta Medical Journal in the article, "The Solvent Action of Papaya Juice on Nitrogenous Articles of Food." In 1879, Papain was named officially by Wurtz and Bouchut, who managed to partially purify the product from the sap of papaya. It wasn't until the mid-twentieth century that the complete purification and isolation of Papain was achieved. In 1968, Drenth et al. determined the structure of Papain by [[X-ray crystallography|x-ray crystallography]], making it the second enzyme whose structure was successfully determined by x-ray crystallography. Additionally, Papain was the first cysteine protease to have its structure identified.<ref name="Worthington" /> In 1984, Kamphuis et al. determined the geometry of the active site, and the three-dimensional structure was visualized to a 1.65 Angstrom solution.<ref name="Structure">PMID:6502713</ref> Today, studies continue on the stability of Papain, involving changes in environmental conditions as well as testing of inhibitors such as phenylmethanesulfonylfluoride (PMSF), TLCK, TPCK, aplh2-macroglobulin, heavy metals, AEBSF, antipain, cystatin, E-64, leupeptin, sulfhydryl binding agents, carbonyl reagents, and alkylating agents.<ref name="Worthington" /> |
| + | |||
| + | Papain is synthesised as an inactive precursor with a '''pro region''' of 107 residues in the N-terminal<ref>PMID:7738022</ref>. | ||
| + | |||
| - | <StructureSection load='9pap' size='500' side='right' caption='Click on the links to the left to view different structural aspects. | ||
| - | PDB code[[9pap]]' scene='Papain/Primary_scene/2' > | ||
==Structure== | ==Structure== | ||
| Line 29: | Line 34: | ||
==Catalytic Mechanism== | ==Catalytic Mechanism== | ||
| + | |||
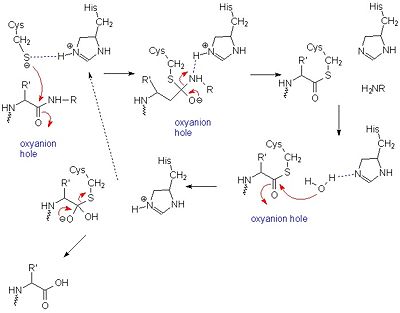
It was once thought that cysteine proteases, like serine proteases, contained a <scene name='Papain/Catalytic_triad/1'>catalytic triad</scene>, consisting of Cys-25, His-159, and Arg-175 <ref> PMID:8140097</ref>. However, site-directed mutagenesis-based studies have demonstrated that Arg-175 is not directly involved in catalysis. Although Arg-175 is clearly important for the enzyme's activity (an Arg175Ala mutation reduces its activity to undetectable levels), this residue neither reacts with the substrate nor modulates the pKa of reacting residues, and therefore cannot be considered catalytic.<ref name="Shokhen">[http://www.ncbi.nlm.nih.gov/pubmed/19688822]Shokhen M, N Khazanov, and A Albeck. 2009. Challenging a paradigm: theoretical calculations of the protonation state of the Cys25-His159 catalytic diad in free papain. Proteins. 77(4):916-26.</ref><ref name="Noble">[http://www.ncbi.nlm.nih.gov/pubmed/11042128]Noble MA, Gul S, Verma CS, Brocklehurst K. 2000. Ionization characteristics and chemical influences of aspartic acid residue 158 of papain and caricain determined by structure-related kinetic and computational techniques: multiple electrostatic modulators of active-centre chemistry. Biochem J. 2000 351: 723-33.</ref> Arg-175 is believed to keep histidine-159 in its stabilized imidazole form, while both histidine-159 and cysteine-25 take part in the actual catalytic mechanism.<ref name="U Maine">[http://chemistry.umeche.maine.edu/CHY431/Peptidase10.html] University of Maine</ref> Despite this, the basic mechanism of papain-catalyzed proteolysis proceeds much like that of serine proteases. The mechanism begins when a peptide binds to the active site. Cys-25 is then deprotonated by His-159 and attacks the substrate carbonyl carbon. This forms a covalent, tetrahedral intermediate that is stabilized by an <scene name='Papain/Oxyanion_hole/1'>oxyanion hole</scene>, formed in large part by Gln-19. Next, His-159 acts as a general acid, protonating the nitrogen in the peptide bond, which acts as a leaving group as the carbonyl reforms. This now free C-terminal portion of the peptide is released. Water then enters the active site and attacks the carbonyl carbon while it is deprotonated by His-159, again forming an oxyanion hole-stabilized tetradral covalent intermediate. Finally, the carbonyl reforms and the Cys-25 sulfur acts as a leaving group, releasing the N-terminal portion of the peptide and regenerating the enzyme. This entire mechanism is shown below<ref name="Harrison">[http://pubs.acs.org/doi/abs/10.1021/ja9711472] Harrison, M.J., N.A. Burton, and I.H. Hillier. 1997. Catalytic Mechanism of the Enzyme Papain: Predictions with a Hybrid Quantum Mechanical/Molecular Mechanical Potential. J. Am. Chem. Soc. 119: 12285-12291</ref>.: | It was once thought that cysteine proteases, like serine proteases, contained a <scene name='Papain/Catalytic_triad/1'>catalytic triad</scene>, consisting of Cys-25, His-159, and Arg-175 <ref> PMID:8140097</ref>. However, site-directed mutagenesis-based studies have demonstrated that Arg-175 is not directly involved in catalysis. Although Arg-175 is clearly important for the enzyme's activity (an Arg175Ala mutation reduces its activity to undetectable levels), this residue neither reacts with the substrate nor modulates the pKa of reacting residues, and therefore cannot be considered catalytic.<ref name="Shokhen">[http://www.ncbi.nlm.nih.gov/pubmed/19688822]Shokhen M, N Khazanov, and A Albeck. 2009. Challenging a paradigm: theoretical calculations of the protonation state of the Cys25-His159 catalytic diad in free papain. Proteins. 77(4):916-26.</ref><ref name="Noble">[http://www.ncbi.nlm.nih.gov/pubmed/11042128]Noble MA, Gul S, Verma CS, Brocklehurst K. 2000. Ionization characteristics and chemical influences of aspartic acid residue 158 of papain and caricain determined by structure-related kinetic and computational techniques: multiple electrostatic modulators of active-centre chemistry. Biochem J. 2000 351: 723-33.</ref> Arg-175 is believed to keep histidine-159 in its stabilized imidazole form, while both histidine-159 and cysteine-25 take part in the actual catalytic mechanism.<ref name="U Maine">[http://chemistry.umeche.maine.edu/CHY431/Peptidase10.html] University of Maine</ref> Despite this, the basic mechanism of papain-catalyzed proteolysis proceeds much like that of serine proteases. The mechanism begins when a peptide binds to the active site. Cys-25 is then deprotonated by His-159 and attacks the substrate carbonyl carbon. This forms a covalent, tetrahedral intermediate that is stabilized by an <scene name='Papain/Oxyanion_hole/1'>oxyanion hole</scene>, formed in large part by Gln-19. Next, His-159 acts as a general acid, protonating the nitrogen in the peptide bond, which acts as a leaving group as the carbonyl reforms. This now free C-terminal portion of the peptide is released. Water then enters the active site and attacks the carbonyl carbon while it is deprotonated by His-159, again forming an oxyanion hole-stabilized tetradral covalent intermediate. Finally, the carbonyl reforms and the Cys-25 sulfur acts as a leaving group, releasing the N-terminal portion of the peptide and regenerating the enzyme. This entire mechanism is shown below<ref name="Harrison">[http://pubs.acs.org/doi/abs/10.1021/ja9711472] Harrison, M.J., N.A. Burton, and I.H. Hillier. 1997. Catalytic Mechanism of the Enzyme Papain: Predictions with a Hybrid Quantum Mechanical/Molecular Mechanical Potential. J. Am. Chem. Soc. 119: 12285-12291</ref>.: | ||
[[Image:Papainmech6.jpg|400px|center|thumb| General mechanism of papain catalysis <ref name="U Maine" />. Arg-175, which orients His 159, and Gln-19, which contributes to the formation of the oxyanion hole, are not shown.]] | [[Image:Papainmech6.jpg|400px|center|thumb| General mechanism of papain catalysis <ref name="U Maine" />. Arg-175, which orients His 159, and Gln-19, which contributes to the formation of the oxyanion hole, are not shown.]] | ||
| Line 57: | Line 63: | ||
There are a small number of <scene name='Papain/Stefin_b_hbonds/2'>direct hydrogen bonds</scene> (labeled in <scene name='Papain/Stefin_b_hbonds/1'>this scene</scene>, between Stefin B and Papain, however there are many more polar interactions mediated by <scene name='Papain/Stfn_b_solvent_intrxns/1'>solvent bridges</scene>, the solvent being mainly <big><b><font color='darkturquoise'>water</font></b></big>. Thirteen solvent molecules of water bridge polar residues of the enzyme and inhibitor. Seventeen hydrogen bonds are made with a solvent molecule and Stefin B. Fourteen of these bridges form a Papain contact. The rest of the interactions are largely hydrophobic-- involving apolar <scene name='Papain/Stefin_b_vdw/2'>Van der Waals forces</scene>.<ref> PMID:2347312 </ref> | There are a small number of <scene name='Papain/Stefin_b_hbonds/2'>direct hydrogen bonds</scene> (labeled in <scene name='Papain/Stefin_b_hbonds/1'>this scene</scene>, between Stefin B and Papain, however there are many more polar interactions mediated by <scene name='Papain/Stfn_b_solvent_intrxns/1'>solvent bridges</scene>, the solvent being mainly <big><b><font color='darkturquoise'>water</font></b></big>. Thirteen solvent molecules of water bridge polar residues of the enzyme and inhibitor. Seventeen hydrogen bonds are made with a solvent molecule and Stefin B. Fourteen of these bridges form a Papain contact. The rest of the interactions are largely hydrophobic-- involving apolar <scene name='Papain/Stefin_b_vdw/2'>Van der Waals forces</scene>.<ref> PMID:2347312 </ref> | ||
| - | |||
| - | __NOTOC__ | ||
| - | </StructureSection> | ||
{{Clear}} | {{Clear}} | ||
| Line 67: | Line 70: | ||
===Medicinal=== | ===Medicinal=== | ||
| - | Papain has been used for a plethora of medicinal purposes including treating inflammation, shingles, diarrhea, psoriasis, parasites, and many others.<ref name="Web MD">[http://www.webmd.com/vitamins-supplements/ingredientmono-69-PAPAIN.aspx?activeIngredientId=69&activeIngredientName=PAPAIN] Web MD </ref> One major use is the treatment of cutaneous ulcers including diabetic ulcers and pressure ulcers. Pressures ulcers plague many bed bound individuals and are a major source of pain and discomfort. Two papain based topical drugs are Accuzyme and Panafil, which can be used to treat wounds like cutaneous ulcers.<ref>[http://www.pbm.va.gov/Clinical%20Guidance/Drug%20Monographs/Papain%20Urea.pdf] National PBM Drug Monograph </ref> | + | Papain has been used for a plethora of medicinal purposes including treating [[Inflammation|inflammation]], shingles, diarrhea, psoriasis, parasites, and many others.<ref name="Web MD">[http://www.webmd.com/vitamins-supplements/ingredientmono-69-PAPAIN.aspx?activeIngredientId=69&activeIngredientName=PAPAIN] Web MD </ref> One major use is the treatment of cutaneous ulcers including diabetic ulcers and pressure ulcers. Pressures ulcers plague many bed bound individuals and are a major source of pain and discomfort. Two papain based topical drugs are Accuzyme and Panafil, which can be used to treat wounds like cutaneous ulcers.<ref>[http://www.pbm.va.gov/Clinical%20Guidance/Drug%20Monographs/Papain%20Urea.pdf] National PBM Drug Monograph </ref> |
A recent New York Times article featured papain and other digestive enzymes. With the number of individuals suffering from irritable bowel syndrome and other gastrointestinal issues, many people are turning toward natural digestive aid supplements like papain. The author even talks about the use of papain along with a pineapple enzyme, bromelain, in cosmetic facial masks. Dr. Adam R. Kolker (a plastic surgeon) is quoted in the article saying that "For skin that is sensitive, enzymes are wonderful." He bases these claims off the idea that proteases like papain help to break peptide bonds holding dead skin cells to the live skin cells.<ref> [http://www.nytimes.com/2012/02/23/fashion/enzymes-once-sidelined-try-to-grab-the-spotlight.html] Enzymes Try to Grab the Spotlight </ref> | A recent New York Times article featured papain and other digestive enzymes. With the number of individuals suffering from irritable bowel syndrome and other gastrointestinal issues, many people are turning toward natural digestive aid supplements like papain. The author even talks about the use of papain along with a pineapple enzyme, bromelain, in cosmetic facial masks. Dr. Adam R. Kolker (a plastic surgeon) is quoted in the article saying that "For skin that is sensitive, enzymes are wonderful." He bases these claims off the idea that proteases like papain help to break peptide bonds holding dead skin cells to the live skin cells.<ref> [http://www.nytimes.com/2012/02/23/fashion/enzymes-once-sidelined-try-to-grab-the-spotlight.html] Enzymes Try to Grab the Spotlight </ref> | ||
===Commercial and Biomedical=== | ===Commercial and Biomedical=== | ||
| + | |||
Papain digests most proteins, often more extensively than pancreatic proteases. It has a very broad specificity and is known to cleave peptide bonds of basic amino acids and leucine and glycine residues, but prefers amino acids with large hydrophobic side chains. This non-specific nature of papain's hydrolase activity has led to its use in many and varied commercial products. It is often used as a meat tenderizer because it can hydrolyze the peptide bonds of collagen, elastin, and actomyosin. It is also used in contact lens solution to remove protein deposits on the lenses and marketed as a digestive supplement. <ref name="Web MD"> Finally, papain has several common uses in general biomedical research, including a gentle cell isolation agent, production of glycopeptides from purified proteoglycans, and solubilization of integral membrane proteins. It is also notable for its ability to specifically cleave IgG and IgM antibodies above and below the disulfide bonds that join the heavy chains and that is found between the light chain and heavy chain. This generates two monovalent Fab segments, that each have a single antibody binding sites, and an intact Fc fragment.<ref name="Worthington" /> | Papain digests most proteins, often more extensively than pancreatic proteases. It has a very broad specificity and is known to cleave peptide bonds of basic amino acids and leucine and glycine residues, but prefers amino acids with large hydrophobic side chains. This non-specific nature of papain's hydrolase activity has led to its use in many and varied commercial products. It is often used as a meat tenderizer because it can hydrolyze the peptide bonds of collagen, elastin, and actomyosin. It is also used in contact lens solution to remove protein deposits on the lenses and marketed as a digestive supplement. <ref name="Web MD"> Finally, papain has several common uses in general biomedical research, including a gentle cell isolation agent, production of glycopeptides from purified proteoglycans, and solubilization of integral membrane proteins. It is also notable for its ability to specifically cleave IgG and IgM antibodies above and below the disulfide bonds that join the heavy chains and that is found between the light chain and heavy chain. This generates two monovalent Fab segments, that each have a single antibody binding sites, and an intact Fc fragment.<ref name="Worthington" /> | ||
| Line 81: | Line 85: | ||
{{Clear}} | {{Clear}} | ||
| - | == | + | </StructureSection> |
| + | ==3D structures of papain== | ||
| - | + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | |
| - | + | {{#tree:id=OrganizedByTopic|openlevels=0| | |
| - | + | *Papain residues 134-345 | |
| - | + | ||
| - | [[ | + | **[[2pad]], [[9pap]], [[1ppn]], [[3lfy]] – PAP - papaya<br /> |
| + | **[[1pad]], [[4pad]], [[5pad]], [[6pad]], [[1bqi]] – PAP + methyl ketone substrate analog <br /> | ||
| + | **[[1pe6]], [[1pip]], [[1ppp]], [[1bp4]], [[1cvz]], [[6tcx]] – PAP + inhibitor<br /> | ||
| + | **[[1khp]], [[1khq]] - PAP + peptide inhibitor<br /> | ||
| + | **[[1ppd]] – hydroxyethyl-thioPAP<br /> | ||
| + | **[[6h8t]] – PAP + Ru complex<br /> | ||
| - | + | *Papain complex with protein inhibitor | |
| - | [[1stf]] - | + | **[[1pop]] – PAP + leupeptin<br /> |
| + | **[[1stf]] – PAP + stefin B<br /> | ||
| + | **[[2cio]] - PAP + cysteine protease inhibitor<br /> | ||
| + | **[[3e1z]] - PAP + chagasin<br /> | ||
| + | **[[3ima]] - PAP + tarocystatin | ||
| - | + | *Pro-papain residues 27-345 | |
| - | [[ | + | **[[3tnx]], [[3usv]], [[4qrg]], [[4qrv]], [[4qrx]] – PPAP (mutant)<br /> |
| + | |||
| + | }} | ||
| + | |||
| + | ==References== | ||
| + | |||
| + | <references /> | ||
| + | <ref group="xtra">PMID:8140097</ref> | ||
| + | |||
| + | {{Clear}} | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
__NOTOC__ | __NOTOC__ | ||
Current revision
| |||||||||||
3D structures of papain
Updated on 18-July-2024
References
- ↑ [1] Papaya's Nutrition Facts
- ↑ [2] Ameridan International
- ↑ [3] Uniprot
- ↑ 4.0 4.1 [4] 9PAP PDB
- ↑ 5.0 5.1 5.2 [5] Sigma Aldrich
- ↑ 6.0 6.1 6.2 [6] Worthington
- ↑ 7.0 7.1 Kamphuis IG, Kalk KH, Swarte MB, Drenth J. Structure of papain refined at 1.65 A resolution. J Mol Biol. 1984 Oct 25;179(2):233-56. PMID:6502713
- ↑ Vernet T, Berti PJ, de Montigny C, Musil R, Tessier DC, Ménard R, Magny MC, Storer AC, Thomas DY. Processing of the papain precursor. The ionization state of a conserved amino acid motif within the Pro region participates in the regulation of intramolecular processing. J Biol Chem. 1995 May 5;270(18):10838-46. PMID:7738022 doi:10.1074/jbc.270.18.10838
- ↑ [7] RCSB PDB
- ↑ [8] Jane S. Richardson
- ↑ [9] The Structure of Papain
- ↑ Berger A, Schechter I. Mapping the active site of papain with the aid of peptide substrates and inhibitors. Philos Trans R Soc Lond B Biol Sci. 1970 Feb 12;257(813):249-64. PMID:4399049
- ↑ Berger A, Schechter I. Mapping the active site of papain with the aid of peptide substrates and inhibitors. Philos Trans R Soc Lond B Biol Sci. 1970 Feb 12;257(813):249-64. PMID:4399049
- ↑ Wang J, Xiang YF, Lim C. The double catalytic triad, Cys25-His159-Asp158 and Cys25-His159-Asn175, in papain catalysis: role of Asp158 and Asn175. Protein Eng. 1994 Jan;7(1):75-82. PMID:8140097
- ↑ [10]Shokhen M, N Khazanov, and A Albeck. 2009. Challenging a paradigm: theoretical calculations of the protonation state of the Cys25-His159 catalytic diad in free papain. Proteins. 77(4):916-26.
- ↑ [11]Noble MA, Gul S, Verma CS, Brocklehurst K. 2000. Ionization characteristics and chemical influences of aspartic acid residue 158 of papain and caricain determined by structure-related kinetic and computational techniques: multiple electrostatic modulators of active-centre chemistry. Biochem J. 2000 351: 723-33.
- ↑ 17.0 17.1 [12] University of Maine
- ↑ [13] Harrison, M.J., N.A. Burton, and I.H. Hillier. 1997. Catalytic Mechanism of the Enzyme Papain: Predictions with a Hybrid Quantum Mechanical/Molecular Mechanical Potential. J. Am. Chem. Soc. 119: 12285-12291
- ↑ [14] Schröder, E., C. Phillips, E. Garman, K. Harlos, C. Crawford. 1997. X-ray crystallographic structure of a papain-leupeptin complex. FEBS Letters 315: 38-42
- ↑ LaLonde JM, Zhao B, Smith WW, Janson CA, DesJarlais RL, Tomaszek TA, Carr TJ, Thompson SK, Oh HJ, Yamashita DS, Veber DF, Abdel-Meguid SS. Use of papain as a model for the structure-based design of cathepsin K inhibitors: crystal structures of two papain-inhibitor complexes demonstrate binding to S'-subsites. J Med Chem. 1998 Nov 5;41(23):4567-76. PMID:9804696 doi:10.1021/jm980249f
- ↑ Valadares NF, Dellamano M, Soares-Costa A, Henrique-Silva F, Garratt RC. Molecular determinants of improved cathepsin B inhibition by new cystatins obtained by DNA shuffling. BMC Struct Biol. 2010 Sep 30;10:30. PMID:20920298 doi:10.1186/1472-6807-10-30
- ↑ Myers MC, Shah PP, Beavers MP, Napper AD, Diamond SL, Smith AB 3rd, Huryn DM. Design, synthesis, and evaluation of inhibitors of cathepsin L: Exploiting a unique thiocarbazate chemotype. Bioorg Med Chem Lett. 2008 Jun 15;18(12):3646-51. Epub 2008 May 1. PMID:18499453 doi:10.1016/j.bmcl.2008.04.065
- ↑ Tsuge H, Nishimura T, Tada Y, Asao T, Turk D, Turk V, Katunuma N. Inhibition mechanism of cathepsin L-specific inhibitors based on the crystal structure of papain-CLIK148 complex. Biochem Biophys Res Commun. 1999 Dec 20;266(2):411-6. PMID:10600517 doi:10.1006/bbrc.1999.1830
- ↑ Beavers MP, Myers MC, Shah PP, Purvis JE, Diamond SL, Cooperman BS, Huryn DM, Smith AB 3rd. Molecular docking of cathepsin L inhibitors in the binding site of papain. J Chem Inf Model. 2008 Jul;48(7):1464-72. Epub 2008 Jul 4. PMID:18598021 doi:10.1021/ci800085c
- ↑ Stubbs MT, Laber B, Bode W, Huber R, Jerala R, Lenarcic B, Turk V. The refined 2.4 A X-ray crystal structure of recombinant human stefin B in complex with the cysteine proteinase papain: a novel type of proteinase inhibitor interaction. EMBO J. 1990 Jun;9(6):1939-47. PMID:2347312
- ↑ [15] RX
- ↑ 27.0 27.1 [16] Web MD
- ↑ [17] National PBM Drug Monograph
- ↑ [18] Enzymes Try to Grab the Spotlight
- ↑ Skern T, Fita I, Guarne A. A structural model of picornavirus leader proteinases based on papain and bleomycin hydrolase. J Gen Virol. 1998 Feb;79 ( Pt 2):301-7. PMID:9472614
Proteopedia Page Contributors and Editors (what is this?)
Kirsten Eldredge, Jacinth Koh, Sara Kongkatong, Kyle Burch, Michal Harel, Joel L. Sussman, Elizabeth Miller, Samuel Bray, David Canner, Jaime Prilusky