P53-DNA Recognition
From Proteopedia
(→Major Groove Base Readout) |
|||
| (10 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| + | {{BAMBED | ||
| + | |DATE=July 26, 2012 | ||
| + | |OLDID=1472467 | ||
| + | |BAMBEDDOI=10.1002/bmb.20650 | ||
| + | }} | ||
| + | <StructureSection load='' size='400' side='right' ' oldscene='Sandbox_Reserved_170/Complex/6' scene='P53-DNA_Recognition/P53_complex/1' caption='Human p53 core complex with DNA (PDB code [[3kz8]]).'> | ||
''This is a joint project of students at La Cañada High School, La Cañada Flintridge, California USA, and students at the University of Southern California, Los Angeles, California USA, mentored by [[User:Remo Rohs|Professor Remo Rohs]].'' | ''This is a joint project of students at La Cañada High School, La Cañada Flintridge, California USA, and students at the University of Southern California, Los Angeles, California USA, mentored by [[User:Remo Rohs|Professor Remo Rohs]].'' | ||
| Line 5: | Line 11: | ||
==Introduction and Biological Role of the Tumor Suppressor p53== | ==Introduction and Biological Role of the Tumor Suppressor p53== | ||
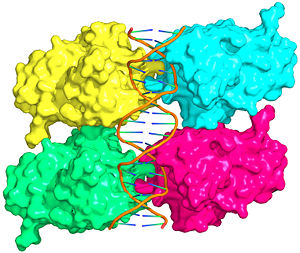
| - | [[Image:p53-intro.jpg|thumb|left|300px|Figure 1: Crystal structure of a p53 DBD tetramer-DNA complex; [ | + | [[Image:p53-intro.jpg|thumb|left|300px|Figure 1: Crystal structure of a p53 DBD tetramer-DNA complex; [[3kz8|PDB ID# 3KZ8]]<ref name='kitayner'>Kitayner M, Rozenberg H, Rohs R, Suad O, Rabinovich D, Honig B, Shakked Z. Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs. Nat Struct Mol Biol. 2010;17(4):423-9. [http://www.ncbi.nlm.nih.gov/pubmed/20364130 PMID:20364130].</ref>.]] |
| - | + | {{Clear}} | |
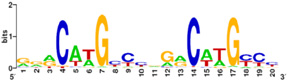
| - | [[Image:consensus.jpg|thumb| | + | [[Image:consensus.jpg|thumb|left|250px|Figure 2: p53 consensus site; R= A or G, Y= C or T, and W=A or T.]] |
| - | + | {{Clear}} | |
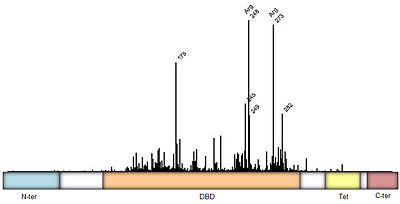
| - | [[Image:p53-domains.jpg|thumb| | + | [[Image:p53-domains.jpg|thumb|left|400px|Figure 3: Frequency of p53 mutants associated with cancer derived from [http://www-p53.iarc.fr/ IARC TP53 database]. Domain architecture; N-ter=N-terminal, DBD=DNA binding domain<ref name='kitayner'/>, Tet=Tetramerization<ref name='tetra'>Jeffrey PD, Gorina S, Pavletich NP. Crystal structure of the p53 tetramerization domain. Science 1995;267:1498-502. [http://www.ncbi.nlm.nih.gov/pubmed/7878469 PMID:7878469].</ref>, and C-ter=C-terminal domain. Intermediate regions are fairly disordered.]] |
| - | + | {{Clear}} | |
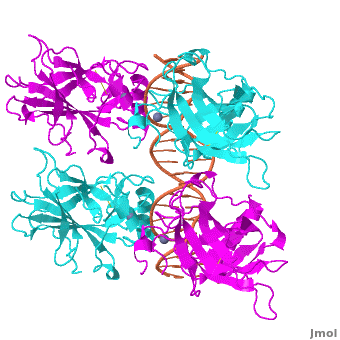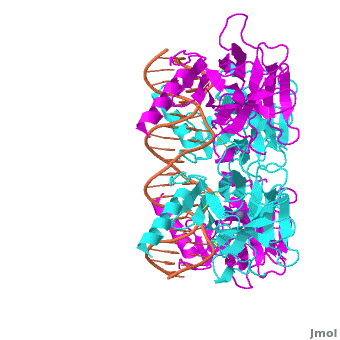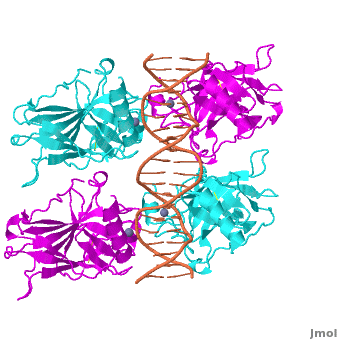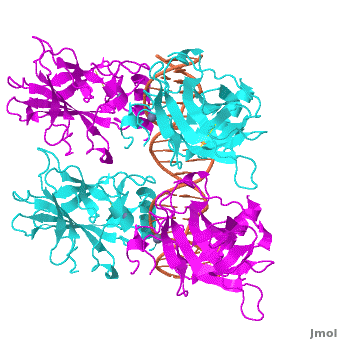
Also known as the '''Guardian of the Genome''', the tumor suppressor p53 is crucial in the natural defense against human cancer. The protein is activated by stress factors that can compromise the genomic integrity of the cell. This activation unleashes the function of p53 as a [[transcription factor]]. It binds as a tetramer ('''Figure 1''') to a large range of DNA response elements. The p53 consensus site ('''Figure 2''') is formed by two decameric half-sites, each containing a core element (red), that are separated by a variable number of base pairs (blue). | Also known as the '''Guardian of the Genome''', the tumor suppressor p53 is crucial in the natural defense against human cancer. The protein is activated by stress factors that can compromise the genomic integrity of the cell. This activation unleashes the function of p53 as a [[transcription factor]]. It binds as a tetramer ('''Figure 1''') to a large range of DNA response elements. The p53 consensus site ('''Figure 2''') is formed by two decameric half-sites, each containing a core element (red), that are separated by a variable number of base pairs (blue). | ||
| Line 21: | Line 27: | ||
===Domain Architecture and Tetramerization=== | ===Domain Architecture and Tetramerization=== | ||
| - | The p53 protein consists of the N-terminal transactivation domain, the DNA binding domain ('''DBD''') or core, the tetramerization domain ([[#Tetramerization Domain|see its structure below]]), and the C-terminal regulatory domain ('''Figure 3'''). This Proteopedia page discusses protein-DNA recognition by p53, thus focusing on the DBD of p53 ('''Figure 4 | + | The p53 protein consists of the N-terminal transactivation domain, the DNA binding domain ('''DBD''') or core, the tetramerization domain ([[#Tetramerization Domain|see its structure below]]), and the C-terminal regulatory domain ('''Figure 3'''). This Proteopedia page discusses protein-DNA recognition by p53, thus focusing on the DBD of p53 (<scene oldname='Sandbox_Reserved_170/Complex/6' name='P53-DNA_Recognition/P53_complex/1'>Figure 4: Crystal structure of p53 DBD tetramer-DNA complex</scene>, [[3kz8|PDB ID 3KZ8]]). |
| - | + | The DBD in tetrameric form binds to a <font color="#e06000">'''DNA response element'''</font>, which consists of two DNA half sites. These decameric half sites can be separated by a DNA spacer of flexible length but in this case, the spacer is of length zero base pairs. The <scene oldname='Sandbox_Reserved_170/Complex/7' name='P53-DNA_Recognition/P53_complex/2'>p53 tetramer binds DNA as a dimer of dimers</scene> with each <font color='e000e0'>'''magenta'''</font>-<font color='00c0c0'>'''cyan'''</font> dimer binding to one half site of the response element<ref>Kitayner M, Rozenberg H, Kessler N, Rabinovich D, Shaulov L, Haran TE, Shakked Z. Structural basis of DNA recognition by p53 tetramers. Mol Cell. 2006 Jun 23;22(6):741-53. [http://www.ncbi.nlm.nih.gov/pubmed/16793544 PMID:16793544].</ref>. | |
| - | + | ||
| - | The DBD in tetrameric form binds to a <font color="#e06000">'''DNA response element'''</font> | + | |
The p53 DBD assumes the conformation of an <scene name='Sandbox_Reserved_170/Beta/1'>immunoglobulin-like fold consisting of a beta sandwich</scene>, which binds the response element in the major groove. A functionally important <scene name='Sandbox_Reserved_170/Zn/1'>Zn2+ ion coordinates the Cys176, His179, Cys238, Cys242 residues</scene> and, thus, stabilizes the fold of the DBD. | The p53 DBD assumes the conformation of an <scene name='Sandbox_Reserved_170/Beta/1'>immunoglobulin-like fold consisting of a beta sandwich</scene>, which binds the response element in the major groove. A functionally important <scene name='Sandbox_Reserved_170/Zn/1'>Zn2+ ion coordinates the Cys176, His179, Cys238, Cys242 residues</scene> and, thus, stabilizes the fold of the DBD. | ||
| Line 36: | Line 40: | ||
[[Image:p53-motif.jpg|thumb|right|300px|Figure 5: p53 binding site motif with G/C base pairs most conserved. PLoS has provided permission for usage of this figure<ref>Horvath MM, Wang X, Resnick MA, Bell DA. Divergent evolution of human p53 binding sites: cell cycle versus apoptosis. PLoS Genet. 2007 Jul;3(7):e127. [http://www.ncbi.nlm.nih.gov/pubmed/17677004 PMID:17677004].</ref>.]] | [[Image:p53-motif.jpg|thumb|right|300px|Figure 5: p53 binding site motif with G/C base pairs most conserved. PLoS has provided permission for usage of this figure<ref>Horvath MM, Wang X, Resnick MA, Bell DA. Divergent evolution of human p53 binding sites: cell cycle versus apoptosis. PLoS Genet. 2007 Jul;3(7):e127. [http://www.ncbi.nlm.nih.gov/pubmed/17677004 PMID:17677004].</ref>.]] | ||
| - | + | {{Clear}} | |
Protein side chains and base pairs form direct contacts in the major groove. Among which, the <scene name='Sandbox_Reserved_170/Arg280_contact/5'>contact between Arg280 and the guanine of the core element</scene> contributes most to binding specificity. This highly specific readout is due to the <scene oldname='Sandbox_Reserved_170/Arg280_contact/4' name='P53-DNA_Recognition/P53_arg280_contact/1'>bidentate hydrogen bond formed between Arg280 and guanine</scene>. As a result of this '''base readout''' the G/C base pairs in the CWWG core elements are the most conserved positions in p53 response elements ('''Figure 5'''). | Protein side chains and base pairs form direct contacts in the major groove. Among which, the <scene name='Sandbox_Reserved_170/Arg280_contact/5'>contact between Arg280 and the guanine of the core element</scene> contributes most to binding specificity. This highly specific readout is due to the <scene oldname='Sandbox_Reserved_170/Arg280_contact/4' name='P53-DNA_Recognition/P53_arg280_contact/1'>bidentate hydrogen bond formed between Arg280 and guanine</scene>. As a result of this '''base readout''' the G/C base pairs in the CWWG core elements are the most conserved positions in p53 response elements ('''Figure 5'''). | ||
| Line 46: | Line 50: | ||
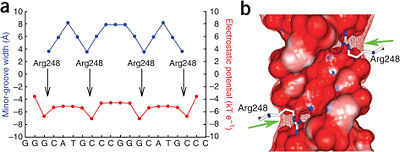
[[Image:Kitayner-etal-Figure7.jpg|thumb|right|400px|Figure 6: DNA shape readout of narrow minor groove regions with enhanced electrostatic potential by Arg248. Nature Publishing Group has provided permission for usage of this figure<ref name='kitayner'/>.]] | [[Image:Kitayner-etal-Figure7.jpg|thumb|right|400px|Figure 6: DNA shape readout of narrow minor groove regions with enhanced electrostatic potential by Arg248. Nature Publishing Group has provided permission for usage of this figure<ref name='kitayner'/>.]] | ||
| - | + | {{Clear}} | |
===Minor Groove Shape Readout=== | ===Minor Groove Shape Readout=== | ||
| Line 58: | Line 62: | ||
==Tetramerization Domain== | ==Tetramerization Domain== | ||
| - | <Structure load='P53tetra.pdb.zip' size='250' frame='true' align='right' caption='Figure 7: Crystal structure of p53 tetramerization domain, [http://proteopedia.com/wiki/index.php/1c26 PDB ID 1C26].' scene='Sandbox_Reserved_170/Tetra/2' /> | ||
| - | Aside from the DBD, the only other domain for which structural information is available is the ''tetramerization domain'' [ | + | Aside from the DBD, the only other domain for which structural information is available is the ''tetramerization domain'' [<scene name='Sandbox_Reserved_170/Tetra/2'>Figure 7: Crystal structure of p53 tetramerization domain</scene>, ([[1c26|PDB ID 1C26]])], which forms as a dimer of dimers with one alpha helix and one beta strand contributed by each p53 monomer. The tetramerization domain is ''not present'' in the crystal structure of the DBD (<scene oldname='Sandbox_Reserved_170/Complex/6' name='P53-DNA_Recognition/P53_complex/1'>Figure 4: Crystal structure of p53 DBD tetramer-DNA complex</scene>). |
=Further Reading= | =Further Reading= | ||
| Line 76: | Line 79: | ||
A more general discussion of structural origins of binding specificity in protein-DNA recognition has been published along with a suggestion for a new '''classification of protein-DNA readout modes''' that goes beyond the historical description of direct and indirect readout<ref name="annualreview">Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. Origins of specificity in protein-DNA recognition. Annu Rev Biochem. 2010;79:233-69. [http://www.ncbi.nlm.nih.gov/pubmed/20334529 PMID:20334529].</ref>.<br/> | A more general discussion of structural origins of binding specificity in protein-DNA recognition has been published along with a suggestion for a new '''classification of protein-DNA readout modes''' that goes beyond the historical description of direct and indirect readout<ref name="annualreview">Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. Origins of specificity in protein-DNA recognition. Annu Rev Biochem. 2010;79:233-69. [http://www.ncbi.nlm.nih.gov/pubmed/20334529 PMID:20334529].</ref>.<br/> | ||
| + | </StructureSection> | ||
| + | =3D structures of p53= | ||
| + | [[P53]] | ||
=Acknowledgements= | =Acknowledgements= | ||
| Line 82: | Line 88: | ||
=References= | =References= | ||
<references/> | <references/> | ||
| + | |||
| + | [[Category:Featured in BAMBED]] | ||
Current revision
This page, as it appeared on July 26, 2012, was featured in this article in the journal Biochemistry and Molecular Biology Education.
| |||||||||||
3D structures of p53
Acknowledgements
This Proteopedia page originates from the partnership of the Rohs Laboratory at the University of Southern California with La Cañada High School. This partnership was initiated by Remo Rohs and Patty Compeau in September 2011 as Bioinformatics Institute, which is part of the Institutes of the 21st Century. Advice and technical help by Proteopedia editors Eran Hodis, Eric Martz, Jaime Prilusky, and Joel Sussman is acknowledged.
References
- ↑ 1.0 1.1 1.2 1.3 Kitayner M, Rozenberg H, Rohs R, Suad O, Rabinovich D, Honig B, Shakked Z. Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs. Nat Struct Mol Biol. 2010;17(4):423-9. PMID:20364130.
- ↑ Jeffrey PD, Gorina S, Pavletich NP. Crystal structure of the p53 tetramerization domain. Science 1995;267:1498-502. PMID:7878469.
- ↑ Kitayner M, Rozenberg H, Kessler N, Rabinovich D, Shaulov L, Haran TE, Shakked Z. Structural basis of DNA recognition by p53 tetramers. Mol Cell. 2006 Jun 23;22(6):741-53. PMID:16793544.
- ↑ Horvath MM, Wang X, Resnick MA, Bell DA. Divergent evolution of human p53 binding sites: cell cycle versus apoptosis. PLoS Genet. 2007 Jul;3(7):e127. PMID:17677004.
- ↑ Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461(7268):1248-53. PMID:19865164.
- ↑ Aishima J, Gitti RK, Noah JE, Gan HH, Schlick T, Wolberger C. A Hoogsteen base pair embedded in undistorted B-DNA. Nucleic Acids Res. 2002;30(23):5244-52. PMID:12466549.
- ↑ Chen Y, Dey R, Chen L. Crystal structure of the p53 core domain bound to a full consensus site as a self-assembled tetramer. Structure. 2010;18(2):246-56. PMID:20159469.
- ↑ Nikolova EN, Kim E, Wise AA, O'Brien PJ, Andricioaei I, Al-Hashimi HM. Transient Hoogsteen base pairs in canonical duplex DNA. Nature. 2011;470(7335):498-502. PMID:21270796.
- ↑ Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. Origins of specificity in protein-DNA recognition. Annu Rev Biochem. 2010;79:233-69. PMID:20334529.
Proteopedia Page Contributors and Editors (what is this?)
Remo Rohs, Eric Martz, Alexander Berchansky, Julia Tam, Sharon Kim, Bailey Holmes, Angel Herraez, Joseph M. Steinberger, Eran Hodis, Masha Karelina, Michal Harel, Ana Carolina Dantas Machado, Jaime Prilusky, Skyler Saleebyan, Joel L. Sussman, Keziah Kim