Sandbox Reserved 654
From Proteopedia
| (47 intermediate revisions not shown.) | |||
| Line 5: | Line 5: | ||
| - | = | + | = '''Bromodomain (PCAF)''' = |
| - | <Structure load='1JM4' size='500' frame='true' align='right' caption=' | + | ---- |
| + | == '''Introduction''' == | ||
| + | <Structure load='1JM4' size='500' frame='true' align='right' caption='PCAF bromodomain bound to HIV-1 Tat NMR structure' /> | ||
| - | + | The bromodomain is an evolutionary conserved motif found in chromatin remodeling complexes. It has been identified in over 100 proteins from multiple organisms ranging from unicellular microscopic eukaryotes (e.g., yeast) to humans. The motif is best known for the groundbreaking recent discoveries made to identify it as the only acetyl-lysine binding domain<ref name=a> Dhalluin, C. et al (1999) Nature 399, 491 [http://www.nature.com/nature/journal/v399/n6735/abs/399491a0.html]</ref> and to reveal its highly specific ligand selectivity properties<ref> Zeng, L. (2002) FEBS 513:1, 124 [http://www.ncbi.nlm.nih.gov/pubmed/11911891]</ref>. Due to these novel characteristics, bromodomains are typically found in proteins responsible for modifications in chromatin structure and the regulation of gene expression, such as histone acetyltransferases (HATs), and the ATPase subunits of chromatin remodeling complexes. There are several families of proteins with bromodomains. One such family is the human transcriptional coactivator p300/CBP-associated factor (PCAF) bromodomain. | |
| - | == '''Structure''' == | + | == '''Structure and Function''' == |
| - | Structure stuff<ref> sample ref </ref> | ||
| - | + | == Structure == | |
| - | < | + | |
| - | + | The bromodomain was originally identified as a sequence of roughly 60 amino acid residues that forms 2 alpha helices<ref>Haynes, S.R. et al (1992) Nucleic Acids Res. 20, 2603 [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC312404/]</ref>. However, it is now known that the bromodomain consist of a highly conserved sequence of approximately 110 amino acids<ref> Owen, D. J. et al. (2000) EMBO J. 19(22), 6141 [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC305837/]</ref>. The structure of the PCAF bromodomain consists of a <scene name='Sandbox_Reserved_654/Four-helix/1'>four-helix</scene> bundle (alphaZ, alphaA, alphaB, and alphaC) with a left-handed twist, and a long intervening loop between helices Z and A (ZA loop)<ref name=a/>. The ZA loop of the bromodomain has a defined conformation and is packed against the loop between helices B and C (BC loop) to form a <scene name='Sandbox_Reserved_654/Kac50_pocket/2'>hydrophobic pocket</scene>. This pocket created by the ZA and BC loops is lined by specific residues (Val 752, Ala 757, Tyr 760, Val 763, Tyr 802 and Tyr 809) that support protein-protein interactions. The ZA loop varies in length between different bromodomains, but almost always contains residues corresponding to Phe 748, Pro 751, Pro 758, Tyr 760 and Pro 767<ref name=a/>. | |
| - | The | + | |
| - | <scene name=' | + | |
| - | + | == Function == | |
| - | + | Until recently, the function of the bromodomain remained unknown. Its structure and modularity, along with its feature of both N and C termini located together on one end of the protein, suggested that it played a role in protein-protein interactions. It has now been shown that the hydrophobic pocket formed by the loops is the primary binding site for acetyl-lysine proteins, making the bromodomain a functional site for recognition of acetylated lysine residues that play a role in gene regulation via protein-protein interactions<ref name=a/>. These interactions have been shown via localization and chemical shift experiments that revealed the specific manner with which the bromodomain hydrophobic cavity binds to acetylated lysine residues. | |
| - | + | Once the acetyl-lysine residue makes the initial binding inside the hydrophobic pocket, the ZA and BC loop residues at the entrance of the pocket interact with the amino acids adjacent (+/- 1 or 2) to the already bound acetyl-lysine. Those interactions reinforce binding of the target sequence<ref>Mujtaba, S. et al (2007) Oncogene 26, 5521 [http://www.nature.com/onc/journal/v26/n37/abs/1210618a.html]</ref>. Small structural changes in the conformation of the ZA and BC loops result in exposing other residues that are originally buried within the protein to aid in peptide recognition<ref name=aa> Mujtaba, S. et al (2002) Mol. Cell 9, 575 [http://www.cell.com/molecular-cell/retrieve/pii/S1097276502004835]</ref>. | |
| - | + | It is also believed that the bromodomain may also play a role in highly specific histone acetylation by tethering transcriptional HATs to specific chromosomal sites<ref> Brownell, J. et al (1996) Curr. Opin. Genet. Dev. 6, 176 [http://www.sciencedirect.com/science/article/pii/S0959437X96800487]</ref> as well as the assembly of multiprotein complexes in transcriptional activation such as the Bromodomain–HIV-1 Tat complex necessary for HIV-1 transcriptional activation<ref name=aa/>. | |
== '''Mechanism''' == | == '''Mechanism''' == | ||
| - | [[Image:F1.medium.gif |thumb|250 px|right| Acetylation of the lysine and its effects on chromatin remodeling..]] | ||
| - | The mechanism of protein-protein interaction for the bromodomain of PCAF with histone proteins begins with the acetylation of lysine residues on the histone tail. The acetylation causes a conformational change in the histones, which allows for transcriptional machinery to access DNA. The bromodomains of PCAF within a pretranscriptional initiation complex (PIC) bind to the acetyl-lysine of the histone to stabilize the complex so that transcription may begin. The bromodomains of PCAF have three major points of contact that allow for site-specific histone recognition. First, the acetylated lysine of the target protein enters a hydrophobic pocket embedded between the ZA and BC loops at the bottom of the protein. The Asn803 residue in the bromodomain forms a hydrogen bond with the amide nitrogen of the acetyl-lysine. Next, residues in the ZA and/or BC loops interact with residues adjacent to the acetyl-lysine, which reinforces the acetyl-lysine binding in the bromodomain. Finally, additional residues in the ZA and BC loops that face opposite to the bromodomain form hydrophobic and/ or electrostatic interaction with the target protein 3 residues away from the acetyl-lysine. This residue clamps on the BC loop together with the acetyl-lysine side chain that is bound inside the hydrophobic pocket of the bromodomain.[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3339198/] | ||
| + | [[Image:F1.medium.gif |thumb|250 px|right| Acetylation of the lysine and its effects on chromatin remodeling.]] | ||
| + | The mechanism of protein-protein interaction for the bromodomain of PCAF with target proteins, such as histones<ref name =aaa>Zeng, L. et al (2008) Structural basis of site-specific histone recognition by the bromodomains of human coactivators PCAF and CBP/p300. Structure 16: 643–652 [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3339198/]</ref> and Tat<ref name=aa/>, begins with the acetylation of lysine residues. The acetylation causes a conformational changes in the histones, which allows for transcriptional machinery to access DNA. The bromodomains of PCAF have three major points of contact that allow for site-specific histone recognition. First, the <scene name='Sandbox_Reserved_654/Kac50/2'>acetylated lysine</scene> of the target protein enters a <scene name='Sandbox_Reserved_654/Kac50_pocket/2'>hydrophobic pocket</scene> embedded between the ZA and BC loops at the bottom of the protein. The Asn803 residue in the bromodomain forms a hydrogen bond with the amide nitrogen of the acetyl-lysine. Next, residues in the ZA and/or BC loops interact with residues adjacent to the acetyl-lysine, which reinforces the acetyl-lysine binding in the bromodomain. Finally, additional residues in the ZA and BC loops that face opposite to the bromodomain form hydrophobic and/ or electrostatic interaction with the target protein 3 residues away from the acetyl-lysine. This residue clamps on the BC loop together with the acetyl-lysine side chain that is bound inside the hydrophobic pocket of the bromodomain<ref name =aaa/>. | ||
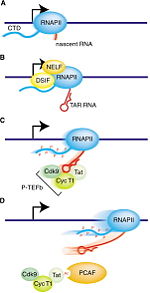
| - | The histone acetyltransferase portion of PCAF helps with the transactivation of HIV-1 by acetylating Lys28 of Tat. The acetylated Lys28 of Tat interacts with positive elongation factors, which stimulates elongation of nascent HIV-1 transcripts. [http://www.sciencedirect.com/science/article/pii/S0969212602007542] | ||
| + | The histone acetyltransferase portion of PCAF helps with the transactivation of HIV-1 by acetylating Lys28 of Tat. The acetylated Lys28 of Tat interacts with positive elongation factors, which stimulates elongation of nascent HIV-1 transcripts. Acetylated Lys50 on Tat causes dissociation from TAR RNA and binds to the bromodomain of PCAF<ref>Nakatani, Y. (2002) HIV-1 Transcription: Activation Mediated by Acetylation of Tat. Structure 10:443-444 | ||
| + | [http://www.sciencedirect.com/science/article/pii/S0969212602007542]</ref>. | ||
| - | + | == '''PCAF Bromodomain-HIV-1 Tat Interaction and Implications''' == | |
| - | == ''' | + | == Interaction and Implications == |
| - | + | <Structure load='1WUG' size='300' frame='true' align='left' caption='PCAF Bromodomain bound to a small ligand molecule designed to inhibit Tat/PCAF association' scene='Insert optional scene name here' /> | |
| + | [[Image:1-s2.0-S0969212602007542-gr1.jpg |thumb|150 px|right| Transcriptional elongation by HIV-1 Tat.]] | ||
| - | + | Protein lysine acetylation is a crucial regulatory mechanism in chromatin remodeling and transcription activation for numerous cellular processes. The human immunodeficiency virus type 1 (HIV-1) trans-activator protein (Tat), for example, stimulates transcription of the HIV genome and promotes viral replication in cells. But Tat transactivation activity is dependent on the acetylation of Lys-50 by p300/CBP<ref name=aa/>. When Tat is acetylated at the Lys-50 residue, Tat dissociates from TAR RNA and binds to the PCAF bromodomain instead. This promotes the formation of a multiprotein complex that is responsible for transcription activation of the HIV genome. | |
| - | + | ||
| - | + | ||
| - | + | == Drug Design == | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ==== | + | |
| - | + | ||
| + | Current anti-HIV drugs target viral proteins such as reverse transcriptase, protease, and integrase<ref>Garg, R. et al (1999) Chem. Rev. 99, 3525 [http://pubs.acs.org/doi/abs/10.1021/cr9703358]</ref>. However, the discovery that Tat transactivation requires Lys-50 acetylation for functional transcription of the viral genome reveals a whole new approach to interfering with virus production. Drugs that target viral proteins have proven inadequate in eradicating the virus because the fast rate of mutations in the target proteins lead to developed drug resistance. Targeting a host cell protein that is necessary for viral reproduction (such as the PCAF bromodomain) as opposed to a viral protein, has the potential to inhibit HIV production much more effectively by disrupting HIV gene expression. | ||
| + | Given the high selectivity of the bromodomain for its target protein, <scene name='Sandbox_Reserved_654/Ligand/1'>small ligand molecules</scene> are currently being designed and engineered to block Tat/PCAF association<ref>Zeng, L. (2005) J. Am. Chem. Soc. 127, 2376 [http://www.ncbi.nlm.nih.gov/pubmed/15724976]</ref>. In addition, new functions of the bromodomain remain to be discovered with implications for many human diseases such as cancer and Alzheimer's disease, and well as breakthroughs in our knowledge of transcription and gene regulation. | ||
== '''References''' == | == '''References''' == | ||
<references/> | <references/> | ||
Current revision
| This Sandbox is Reserved from 30/08/2012, through 01/02/2013 for use in the course "Proteins and Molecular Mechanisms" taught by Robert B. Rose at the North Carolina State University, Raleigh, NC USA. This reservation includes Sandbox Reserved 636 through Sandbox Reserved 685. | |||||||||||||
To get started:
More help: Help:Editing For more help, look at this link: http://proteopedia.org/w/Help:Getting_Started_in_Proteopedia
Bromodomain (PCAF)Introduction
The bromodomain is an evolutionary conserved motif found in chromatin remodeling complexes. It has been identified in over 100 proteins from multiple organisms ranging from unicellular microscopic eukaryotes (e.g., yeast) to humans. The motif is best known for the groundbreaking recent discoveries made to identify it as the only acetyl-lysine binding domain[1] and to reveal its highly specific ligand selectivity properties[2]. Due to these novel characteristics, bromodomains are typically found in proteins responsible for modifications in chromatin structure and the regulation of gene expression, such as histone acetyltransferases (HATs), and the ATPase subunits of chromatin remodeling complexes. There are several families of proteins with bromodomains. One such family is the human transcriptional coactivator p300/CBP-associated factor (PCAF) bromodomain. Structure and FunctionStructureThe bromodomain was originally identified as a sequence of roughly 60 amino acid residues that forms 2 alpha helices[3]. However, it is now known that the bromodomain consist of a highly conserved sequence of approximately 110 amino acids[4]. The structure of the PCAF bromodomain consists of a bundle (alphaZ, alphaA, alphaB, and alphaC) with a left-handed twist, and a long intervening loop between helices Z and A (ZA loop)[1]. The ZA loop of the bromodomain has a defined conformation and is packed against the loop between helices B and C (BC loop) to form a . This pocket created by the ZA and BC loops is lined by specific residues (Val 752, Ala 757, Tyr 760, Val 763, Tyr 802 and Tyr 809) that support protein-protein interactions. The ZA loop varies in length between different bromodomains, but almost always contains residues corresponding to Phe 748, Pro 751, Pro 758, Tyr 760 and Pro 767[1]. FunctionUntil recently, the function of the bromodomain remained unknown. Its structure and modularity, along with its feature of both N and C termini located together on one end of the protein, suggested that it played a role in protein-protein interactions. It has now been shown that the hydrophobic pocket formed by the loops is the primary binding site for acetyl-lysine proteins, making the bromodomain a functional site for recognition of acetylated lysine residues that play a role in gene regulation via protein-protein interactions[1]. These interactions have been shown via localization and chemical shift experiments that revealed the specific manner with which the bromodomain hydrophobic cavity binds to acetylated lysine residues. Once the acetyl-lysine residue makes the initial binding inside the hydrophobic pocket, the ZA and BC loop residues at the entrance of the pocket interact with the amino acids adjacent (+/- 1 or 2) to the already bound acetyl-lysine. Those interactions reinforce binding of the target sequence[5]. Small structural changes in the conformation of the ZA and BC loops result in exposing other residues that are originally buried within the protein to aid in peptide recognition[6]. It is also believed that the bromodomain may also play a role in highly specific histone acetylation by tethering transcriptional HATs to specific chromosomal sites[7] as well as the assembly of multiprotein complexes in transcriptional activation such as the Bromodomain–HIV-1 Tat complex necessary for HIV-1 transcriptional activation[6]. MechanismThe mechanism of protein-protein interaction for the bromodomain of PCAF with target proteins, such as histones[8] and Tat[6], begins with the acetylation of lysine residues. The acetylation causes a conformational changes in the histones, which allows for transcriptional machinery to access DNA. The bromodomains of PCAF have three major points of contact that allow for site-specific histone recognition. First, the of the target protein enters a embedded between the ZA and BC loops at the bottom of the protein. The Asn803 residue in the bromodomain forms a hydrogen bond with the amide nitrogen of the acetyl-lysine. Next, residues in the ZA and/or BC loops interact with residues adjacent to the acetyl-lysine, which reinforces the acetyl-lysine binding in the bromodomain. Finally, additional residues in the ZA and BC loops that face opposite to the bromodomain form hydrophobic and/ or electrostatic interaction with the target protein 3 residues away from the acetyl-lysine. This residue clamps on the BC loop together with the acetyl-lysine side chain that is bound inside the hydrophobic pocket of the bromodomain[8].
PCAF Bromodomain-HIV-1 Tat Interaction and ImplicationsInteraction and Implications
Protein lysine acetylation is a crucial regulatory mechanism in chromatin remodeling and transcription activation for numerous cellular processes. The human immunodeficiency virus type 1 (HIV-1) trans-activator protein (Tat), for example, stimulates transcription of the HIV genome and promotes viral replication in cells. But Tat transactivation activity is dependent on the acetylation of Lys-50 by p300/CBP[6]. When Tat is acetylated at the Lys-50 residue, Tat dissociates from TAR RNA and binds to the PCAF bromodomain instead. This promotes the formation of a multiprotein complex that is responsible for transcription activation of the HIV genome. Drug DesignCurrent anti-HIV drugs target viral proteins such as reverse transcriptase, protease, and integrase[10]. However, the discovery that Tat transactivation requires Lys-50 acetylation for functional transcription of the viral genome reveals a whole new approach to interfering with virus production. Drugs that target viral proteins have proven inadequate in eradicating the virus because the fast rate of mutations in the target proteins lead to developed drug resistance. Targeting a host cell protein that is necessary for viral reproduction (such as the PCAF bromodomain) as opposed to a viral protein, has the potential to inhibit HIV production much more effectively by disrupting HIV gene expression. Given the high selectivity of the bromodomain for its target protein, are currently being designed and engineered to block Tat/PCAF association[11]. In addition, new functions of the bromodomain remain to be discovered with implications for many human diseases such as cancer and Alzheimer's disease, and well as breakthroughs in our knowledge of transcription and gene regulation. References
|