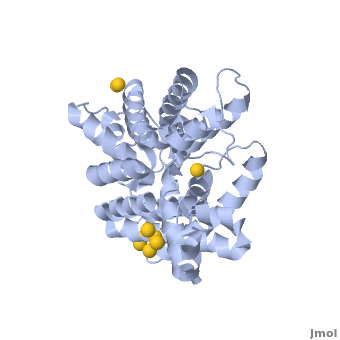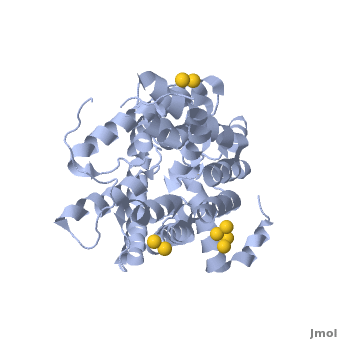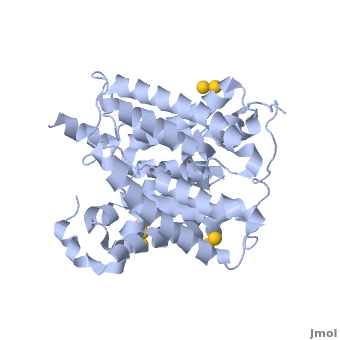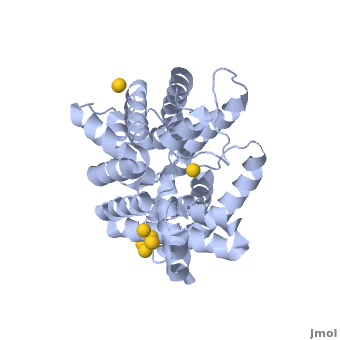
Urea transporter
From Proteopedia
| (12 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | [[ | + | <StructureSection load='' size='350' side='right' caption='Urea transporter ([[3me1]]' scene='Urea_transporter/3k3fdefault/1' pspeed='8'> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
===Crystal Structure of the Urea Transporter from Desulfovibrio Vulgaris=== | ===Crystal Structure of the Urea Transporter from Desulfovibrio Vulgaris=== | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
{{ABSTRACT_PUBMED_19865084}} | {{ABSTRACT_PUBMED_19865084}} | ||
==About this Structure== | ==About this Structure== | ||
| - | {{Structure | ||
| - | |PDB= 3me1 |SIZE=400|SCENE= |CAPTION= urea transporter trimer ([[3me1]]); (<scene name='Urea_transporter/3k3fdefault/1'>initial scene</scene>) | ||
| - | |SITE= | ||
| - | |LIGAND= | ||
| - | |ACTIVITY= | ||
| - | |GENE= | ||
| - | |DOMAIN= | ||
| - | |RELATEDENTRY= | ||
| - | |RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3k3f FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3k3f OCA], [http://www.ebi.ac.uk/pdbsum/3k3f PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=3k3f RCSB]</span> | ||
| - | }} | ||
| - | The structure revealed <scene name='Urea_transporter/3k3fdefault/1a> monomer of the urea transporter</scene> in the asymmetric unit. | ||
| - | However, the urea transporter crystallized as <scene name='Urea_transporter/3k3f_trimer/1'>a homotrimer that corresponds to its functional biological state</scene>, and this is seen when considering the crystallographic three-fold symmetry axis. | ||
| - | To help give a better idea of how the urea transporter is oriented in the membrane lipid bilayer, <scene name='Urea_transporter/Opm_3k3f/3'>a slab representative of hydrophobic core of the lipid bilayer</scene> as calculated by the [http://opm.phar.umich.edu/protein.php?pdbid=3kg2 Orientations of Proteins in Membranes database](University of Michigan, USA) is shown with the red patch of spheres indicating the boundary of the hydrophobic core closet to the outside of the cell and the dark blue patch of spheres indicating the boundary closest to the inside of the cell. | + | To help give a better idea of how the '''urea transporter''' is oriented in the membrane lipid bilayer, <scene name='Urea_transporter/Opm_3k3f/3'>a slab representative of hydrophobic core of the lipid bilayer</scene> as calculated by the [http://opm.phar.umich.edu/protein.php?pdbid=3kg2 Orientations of Proteins in Membranes database](University of Michigan, USA) is shown with the red patch of spheres indicating the boundary of the hydrophobic core closet to the outside of the cell and the dark blue patch of spheres indicating the boundary closest to the inside of the cell. |
[[Image:Opm_periplasmic_topology.gif]] <!--I got this from Eric Martz's [[Mechanosensitive channels: opening and closing]]--> | [[Image:Opm_periplasmic_topology.gif]] <!--I got this from Eric Martz's [[Mechanosensitive channels: opening and closing]]--> | ||
| - | ==3D structures of urea transporter== | + | </StructureSection> |
| + | ==3D structures of '''urea transporter''' or '''solute carrier family 14'''== | ||
| + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | |||
| + | [[8blp]] – hUT 1 + cholesterol derivative – human<br /> | ||
| + | [[6qd5]] – hUT 1 (mutant) + cholesterol derivative <br /> | ||
| + | [[8blo]] – hUT 2 + choline derivative <br /> | ||
| + | [[4ezc]] – bUT + ceramide - bovine<br /> | ||
| + | [[4ezd]] – bUT + ceramide + selenourea<br /> | ||
[[3k3f]], [[3me1]] – DvUT – ''Desulfovibrio vulgaris''<br /> | [[3k3f]], [[3me1]] – DvUT – ''Desulfovibrio vulgaris''<br /> | ||
[[3m6e]] – DvUT (mutant)<br /> | [[3m6e]] – DvUT (mutant)<br /> | ||
| - | [[3k3g]] – DvUT + dimethylurea | + | [[3k3g]] – DvUT + dimethylurea<br /> |
| - | + | [[7s6e]] – UT + urea – ''synechococcus''<br /> | |
==Reference for the structure== | ==Reference for the structure== | ||
| Line 65: | Line 51: | ||
''Page started with original page on [[3k3f]] seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu Jan 28 14:58:46 2010'' | ''Page started with original page on [[3k3f]] seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu Jan 28 14:58:46 2010'' | ||
| + | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
Contents |
3D structures of urea transporter or solute carrier family 14
Updated on 13-March-2024
8blp – hUT 1 + cholesterol derivative – human
6qd5 – hUT 1 (mutant) + cholesterol derivative
8blo – hUT 2 + choline derivative
4ezc – bUT + ceramide - bovine
4ezd – bUT + ceramide + selenourea
3k3f, 3me1 – DvUT – Desulfovibrio vulgaris
3m6e – DvUT (mutant)
3k3g – DvUT + dimethylurea
7s6e – UT + urea – synechococcus
Reference for the structure
- Levin EJ, Quick M, Zhou M. Crystal structure of a bacterial homologue of the kidney urea transporter. Nature. 2009 Dec 10;462(7274):757-61. Epub . PMID:19865084 doi:10.1038/nature08558
See Also
- Knepper MA, Mindell JA. Structural biology: Molecular coin slots for urea. Nature. 2009 Dec 10;462(7274):733-4. PMID:20010678 doi:10.1038/462733a
- For Additional Information, See Membrane Channels & Pumps
Reference
Page started with original page on 3k3f seeded by OCA on Thu Jan 28 14:58:46 2010
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Wayne Decatur, David Canner, Jaime Prilusky, Joel L. Sussman
Categories: Desulfovibrio vulgaris str. hildenborough | Levin, E J. | NYCOMPS, New York Consortium on Membrane Protein Structure. | Zhou, M. | Channel | Membrane protein | New york consortium on membrane protein structure | Nycomp | Protein structure initiative | Psi-2 | Structural genomic | Transport protein | Transporter | Urea transport | Topic Page