Circadian Clock Protein KaiC
From Proteopedia
(→Introduction) |
|||
| (5 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| + | <StructureSection load='1tf7' size='350' side='right' caption='Structure of KaiC complex with ATP (PDB entry [[1tf7]])' scene=''> | ||
==Introduction== | ==Introduction== | ||
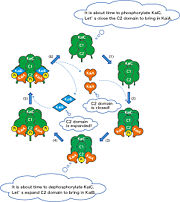
[[Image:KaiCABinteraction.jpg | thumb | alt=text | Interactions of KaiC with KaiA and KaiB ]] | [[Image:KaiCABinteraction.jpg | thumb | alt=text | Interactions of KaiC with KaiA and KaiB ]] | ||
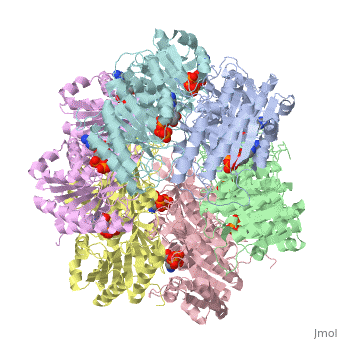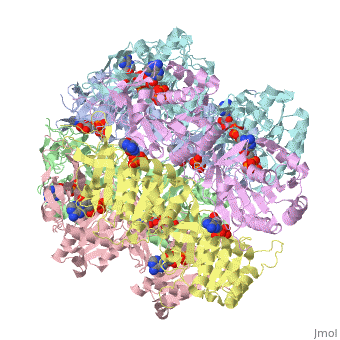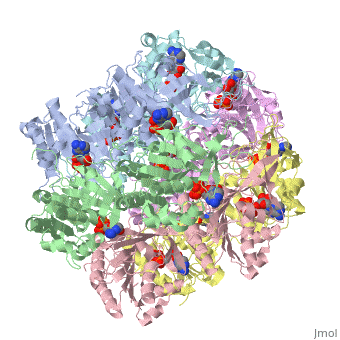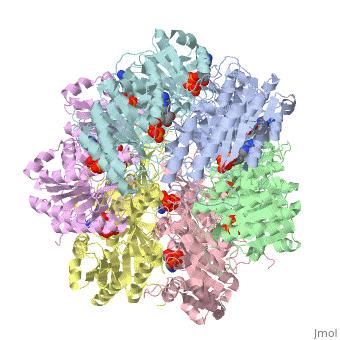
| - | Biological Circadian Clocks are self-sustaining oscillators that function on a rhythmic cycle 24 hours. They exhibit a very precise time constant and temperature compensation, which allows the system to run at a steady rate independent of temperature fluctuations <ref name= Johnson> PMCID: PMC2585598 </ref>. They are found in almost all organisms, the simplest of which are cyanobacteria, which have been extensively studied in order to determine the mechanism of the fine-tuned biological process of circadian rhythmicity. In most eukaryotes, a region of the brain called the superchiasmic nuclei detects light and dark cycles, then relays the message to biological clock systems that maintain rhythmicity within the body. Conversely, cyanobacteria have a fairly modest system comprised of three proteins: KaiC, KaiA, and KaiB. The system is based around the central protein KaiC which exhibits ATP binding, inter-subunit organization, a scaffold region for Kai protein complex formation, a location where critical mutations are found, and an evolutionary link to other well-known proteins <ref name= Pattanayek> PMID: 15304218 </ref>. In order to devise an explanation for the mechanism of biological oscillators, we need to characterize the structure, function, and interactions among molecular components. To study these, scientists begin with analyzing cyanobacteria such as ''Synechococcus elongatus'', since it is the smallest organism that expresses rhythmic clock properties. | + | Biological Circadian Clocks are self-sustaining oscillators that function on a rhythmic cycle 24 hours. They exhibit a very precise time constant and temperature compensation, which allows the system to run at a steady rate independent of temperature fluctuations <ref name= Johnson> PMCID: PMC2585598 </ref>. They are found in almost all organisms, the simplest of which are cyanobacteria, which have been extensively studied in order to determine the mechanism of the fine-tuned biological process of circadian rhythmicity. In most eukaryotes, a region of the brain called the superchiasmic nuclei detects light and dark cycles, then relays the message to biological clock systems that maintain rhythmicity within the body. Conversely, cyanobacteria have a fairly modest system comprised of three proteins: '''KaiC, KaiA, and KaiB'''. The system is based around the central protein KaiC which exhibits ATP binding, inter-subunit organization, a scaffold region for Kai protein complex formation, a location where critical mutations are found, and an evolutionary link to other well-known proteins <ref name= Pattanayek> PMID: 15304218 </ref>. In order to devise an explanation for the mechanism of biological oscillators, we need to characterize the structure, function, and interactions among molecular components. To study these, scientists begin with analyzing cyanobacteria such as ''Synechococcus elongatus'', since it is the smallest organism that expresses rhythmic clock properties. |
==KaiC - KaiA - KaiB System== | ==KaiC - KaiA - KaiB System== | ||
| Line 7: | Line 8: | ||
| - | + | ||
== KaiC Homohexameric Complex == | == KaiC Homohexameric Complex == | ||
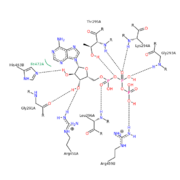
[[Image:ATP-1TF7 psv v 500.png | thumb | alt=text | Intramolecular interactions of ATP binding site ]] | [[Image:ATP-1TF7 psv v 500.png | thumb | alt=text | Intramolecular interactions of ATP binding site ]] | ||
| Line 19: | Line 20: | ||
A region of each hexamer that is notable regarding the phosphorylation of KaiC is the <scene name='Circadian_Clock_Protein_KaiC/P-loop-shuttle-btw-431-432-426/1'>P-Loop</scene>. This zone is recognized as site for binding and hydrolysis of ATP. Along with the T432 site, evidence shows a shuttling of phosphates between residues <scene name='Circadian_Clock_Protein_KaiC/Active_site_t432_t426_s431/1'>S431 and T426</scene> of the P-Loop. | A region of each hexamer that is notable regarding the phosphorylation of KaiC is the <scene name='Circadian_Clock_Protein_KaiC/P-loop-shuttle-btw-431-432-426/1'>P-Loop</scene>. This zone is recognized as site for binding and hydrolysis of ATP. Along with the T432 site, evidence shows a shuttling of phosphates between residues <scene name='Circadian_Clock_Protein_KaiC/Active_site_t432_t426_s431/1'>S431 and T426</scene> of the P-Loop. | ||
| - | Based on RecA structure/function similarity, a conserved glutamate <scene name='Circadian_Clock_Protein_KaiC/P-loop-431-432-426-e318/2'>E318</scene> in CI is proposed to activate water for an attack on the gamma-phosphate of ATP in order to release the phosphate to be shuttled. The <scene name='Circadian_Clock_Protein_KaiC/Active-site-of-a-b/1'>ATP binding</scene> site within the A and B monomer and its interactions are displayed in the image on the | + | Based on RecA structure/function similarity, a conserved glutamate <scene name='Circadian_Clock_Protein_KaiC/P-loop-431-432-426-e318/2'>E318</scene> in CI is proposed to activate water for an attack on the gamma-phosphate of ATP in order to release the phosphate to be shuttled. The <scene name='Circadian_Clock_Protein_KaiC/Active-site-of-a-b/1'>ATP binding</scene> site within the A and B monomer and its interactions are displayed in the image on the right which shows hydrogen bonds and interactions between molecules which hold the ATP in the pocket. |
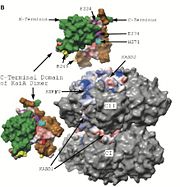
==KaiA - KaiC Interaction Site== | ==KaiA - KaiC Interaction Site== | ||
| Line 37: | Line 38: | ||
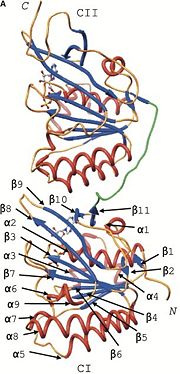
[[Image:ATP-bind-site-cI(A-cropped).jpg | thumb | alt=text | Secondary Structure of Monomers ]] | [[Image:ATP-bind-site-cI(A-cropped).jpg | thumb | alt=text | Secondary Structure of Monomers ]] | ||
In cyanobacteria, the KaiC system is vital to survival because of it's role in global gene regulation: nearly all promoters in a cyanobacteria are under circadian control. Correlating with a circadian clock system enhances fitness of any organism in a rhythmic environment <ref name= Yao> PMCID: PMC518856 </ref>. | In cyanobacteria, the KaiC system is vital to survival because of it's role in global gene regulation: nearly all promoters in a cyanobacteria are under circadian control. Correlating with a circadian clock system enhances fitness of any organism in a rhythmic environment <ref name= Yao> PMCID: PMC518856 </ref>. | ||
| - | Structure similarity exists between KaiC and RecA and DnaB. RecA is a DNA recombinase and DnaB is a DNA helicase, so the observation that there is similarity between these molecules imply possible direct interactions with DNA. The folds of each monomer resemble those of RecA, where eight α-helices surround the twisted β-sheet made up | + | Structure similarity exists between KaiC and RecA and DnaB. RecA is a DNA recombinase and DnaB is a DNA helicase, so the observation that there is similarity between these molecules imply possible direct interactions with DNA. The folds of each monomer resemble those of RecA, where eight α-helices surround the twisted β-sheet made up of seven strands (shown in the figure on the right). The similarity indicates nucleotide binding on the carboxy side of the β-sheet <ref name= Pattanayek> PMID: 15304218</ref>. The entire hexamer also has structural similarities to proteins that do not bind DNA, such as F1-ATPase. This is consistent with the fact that KaiC has phosphotransferase activity, so it is capable of generating ATP. Yet there is more indication that KaiC acts on DNA, especially single stranded DNA, rather than pumps any small molecule through the pore. Inside the CI domain of the channel, the electrostatic potential is mostly negative, while inside the more constricted CII domain of the channel, the electrostatic potential is mostly positive. |
The biological activity of KaiC is still a mystery. It may mediate changes in chromosomal torsion or interact directly with nucleic acids <ref name= Pattanayek> PMID: 15304218</ref>. It has also been shown to cause changes in DNA topology and transcription factor activity <ref name= Johnson> PMCID: PMC2585598 </ref>. Based on the amount of KaiC molecules per cell (~10,000) and its DNA interaction properties, scientists are strongly persuaded that the protein regulates global gene expression by a direct mechanism that changes DNA structure. | The biological activity of KaiC is still a mystery. It may mediate changes in chromosomal torsion or interact directly with nucleic acids <ref name= Pattanayek> PMID: 15304218</ref>. It has also been shown to cause changes in DNA topology and transcription factor activity <ref name= Johnson> PMCID: PMC2585598 </ref>. Based on the amount of KaiC molecules per cell (~10,000) and its DNA interaction properties, scientists are strongly persuaded that the protein regulates global gene expression by a direct mechanism that changes DNA structure. | ||
| - | Defining the mechanism of a biological oscillator is a powerful key to the future. Although biological clock system in bacteria differs vastly from the complex network of molecules that make up eukaryotic biological clocks, it can still be useful to the understanding of circadian rhythms in general. Knowing the mechanism of KaiC can help us eradicate or reduce the harm of certain bacteria by altering their biological rhythm and therefore decreasing their fitness. On the other hand, we can enhance the fitness of other bacteria in order to exploit them for positive changes in the atmosphere, such as bioremedial techniques. The possibilities are endless with such a vital function as circadian rhythms. | + | Defining the mechanism of a biological oscillator is a powerful key to the future. Although biological clock system in bacteria differs vastly from the complex network of molecules that make up eukaryotic biological clocks, it can still be useful to the understanding of circadian rhythms in general. Knowing the mechanism of KaiC can help us eradicate or reduce the harm of certain bacteria by altering their biological rhythm and therefore decreasing their fitness. On the other hand, we can enhance the fitness of other bacteria in order to exploit them for positive changes in the atmosphere, such as bioremedial techniques. The possibilities are endless with such a vital function as circadian rhythms. |
==References== | ==References== | ||
{{Reflist}} | {{Reflist}} | ||
Current revision
| |||||||||||
Biological Importance and Evolutionary Complementarity
In cyanobacteria, the KaiC system is vital to survival because of it's role in global gene regulation: nearly all promoters in a cyanobacteria are under circadian control. Correlating with a circadian clock system enhances fitness of any organism in a rhythmic environment [3]. Structure similarity exists between KaiC and RecA and DnaB. RecA is a DNA recombinase and DnaB is a DNA helicase, so the observation that there is similarity between these molecules imply possible direct interactions with DNA. The folds of each monomer resemble those of RecA, where eight α-helices surround the twisted β-sheet made up of seven strands (shown in the figure on the right). The similarity indicates nucleotide binding on the carboxy side of the β-sheet [2]. The entire hexamer also has structural similarities to proteins that do not bind DNA, such as F1-ATPase. This is consistent with the fact that KaiC has phosphotransferase activity, so it is capable of generating ATP. Yet there is more indication that KaiC acts on DNA, especially single stranded DNA, rather than pumps any small molecule through the pore. Inside the CI domain of the channel, the electrostatic potential is mostly negative, while inside the more constricted CII domain of the channel, the electrostatic potential is mostly positive. The biological activity of KaiC is still a mystery. It may mediate changes in chromosomal torsion or interact directly with nucleic acids [2]. It has also been shown to cause changes in DNA topology and transcription factor activity [1]. Based on the amount of KaiC molecules per cell (~10,000) and its DNA interaction properties, scientists are strongly persuaded that the protein regulates global gene expression by a direct mechanism that changes DNA structure.
Defining the mechanism of a biological oscillator is a powerful key to the future. Although biological clock system in bacteria differs vastly from the complex network of molecules that make up eukaryotic biological clocks, it can still be useful to the understanding of circadian rhythms in general. Knowing the mechanism of KaiC can help us eradicate or reduce the harm of certain bacteria by altering their biological rhythm and therefore decreasing their fitness. On the other hand, we can enhance the fitness of other bacteria in order to exploit them for positive changes in the atmosphere, such as bioremedial techniques. The possibilities are endless with such a vital function as circadian rhythms.
References
- ↑ 1.0 1.1 PMCID: PMC2585598
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Pattanayek R, Wang J, Mori T, Xu Y, Johnson CH, Egli M. Visualizing a circadian clock protein: crystal structure of KaiC and functional insights. Mol Cell. 2004 Aug 13;15(3):375-88. PMID:15304218 doi:10.1016/j.molcel.2004.07.013
- ↑ 3.0 3.1 3.2 PMCID: PMC518856
- ↑ http://www.spring8.or.jp/en/news_publications/publications/scientific_results/life_science/topic3
Proteopedia Page Contributors and Editors (what is this?)
Ashley Beechan, Michal Harel, Alexander Berchansky, Jaime Prilusky