Molecular Playground/Nickel Superoxide Dismutase
From Proteopedia
(Difference between revisions)
| (38 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | == | + | <StructureSection load='1q0m' size='350' side='right' caption='Ni-bound NiSOD (PDB ID: [[1q0m]]). The yellow/red molecules represent sulphates, the grey/red molecules represent acetic acid, and the green spheres represent the nickel ions bound by the nickel-hook region.' scene='' pspeed='8'> |
| - | + | Nickel superoxide dismutase (NiSOD) is one of the [[CBI Molecules]] being studied in the [http://www.umass.edu/cbi/ University of Massachusetts, Amherst Chemistry-Biology Interface Program] at Umass Amherst and on display at the [http://www.molecularplayground.org/ Molecular Playground]. | |
| - | Nickel superoxide dismutase (NiSOD) is one of the [[CBI Molecules]] being studied in the University of Massachusetts, Amherst Chemistry-Biology Interface Program at Umass Amherst and on display at the Molecular Playground. | + | |
===Introduction=== | ===Introduction=== | ||
| + | ---- | ||
| - | Nickel Superoxide Dismutase (NiSOD) is the newest member in a class of enzymes that protects organisms from oxidative stress caused by superoxide, a harmful free radical byproduct of aerobic metabolism. NiSOD reacts with two molecules of superoxide, to form O<sub>2</sub> and H<sub>2</sub>O<sub>2</sub> with rates occurring at or near the diffusion limit. During catalysis, the redox-active nickel center cycles between an oxidized and reduced state. This reaction is termed the | + | '''Nickel Superoxide Dismutase''' (NiSOD) is the newest member in a class of enzymes that protects organisms from oxidative stress caused by superoxide, a harmful free radical byproduct of aerobic metabolism. NiSOD reacts with two molecules of superoxide, to form O<sub>2</sub> and H<sub>2</sub>O<sub>2</sub> with rates occurring at or near the diffusion limit. During catalysis, the redox-active nickel center cycles between an oxidized and reduced state. This reaction is termed the ping pong mechanism and is shown below. |
| - | + | ||
| - | + | ||
:M<sup>(n + 1)</sup> + O<sub>2</sub><sup>•-</sup> → M<sup>n+</sup> + O<sub>2</sub> <br /> | :M<sup>(n + 1)</sup> + O<sub>2</sub><sup>•-</sup> → M<sup>n+</sup> + O<sub>2</sub> <br /> | ||
| Line 15: | Line 13: | ||
NiSOD is unique among SOD's for a variety of reasons. <br /> | NiSOD is unique among SOD's for a variety of reasons. <br /> | ||
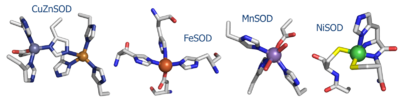
| - | NiSOD shares no sequence homology with the other known SOD's ('''Fig. 1'''). Copper, iron, and manganese SOD are redox active in aqueous media at biological pH. Nickel | + | NiSOD shares no sequence homology with the [http://en.wikipedia.org/wiki/Superoxide_dismutase other known SOD's] ('''Fig. 1''').<sup><ref>PMID: 1094908</ref>-<ref>PMID: 11912934</ref></sup> Copper, iron, and manganese SOD are redox active in aqueous media at biological pH. Nickel is not, and requires the coordination of the two cysteine ligands to tune its redox potential, which is estimated to lie above 2 eV, far outside the biologically relevant redox potential necessary to oxidize or reduce superoxide [-160 to +879 eV].<ref>Uedsemma, M., Tamm, T. Density-functional theory calculations of aqueous redox potentials of fourth-period transition metals. J. Phys. Chem. A. 2003 Aug 4;107:9997-10003. Epub 2003 Oct 23. 10.1021/jp0362741</ref> The ligands employed in the redox-active metal center are distinct. Cu/Zn, Fe, and MnSOD employ only aspartic acids, waters, and histidines.<ref>PMID: 9878406</ref>, <ref>PMID: 1394426</ref>, <ref>PMID: 7849024</ref> In NiSOD, the nickel center is coordinated by the side chains of cysteine 2 and cysteine 6, as well as the N-terminal amine, the amide group of cysteine 2 and an axial histidine ligand ('''Fig. 1'''). <br /> See also [[Superoxide Dismutase]]. |
| - | + | ||
| + | [[Image:SODs.png|400 px|thumb|'''Fig. 1:''' Active site structures of the four known SOD's.]] <br /> | ||
===NiSOD Structure=== | ===NiSOD Structure=== | ||
| + | ---- | ||
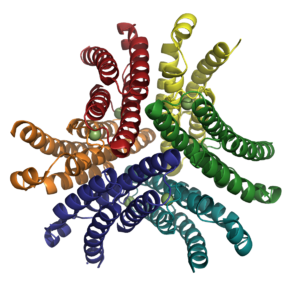
| - | NiSOD is a homohexamer ('''Fig. 2''') composed of a dimer of trimers and binds one nickel ion per monomer. The residues coordinated to the nickel active site are located within the first six N-terminal amino acids, termed the nickel-hook. The nickel in the active site cycles between Ni(II) and Ni(III). In the Ni(II) state the nickel has a square | + | NiSOD is a homohexamer ('''Fig. 2''') composed of a <scene name='User:Carolyn_Carr/Sandbox_1/Nisod_dimer/1'>dimer</scene> of <scene name='User:Carolyn_Carr/Sandbox_1/Nisod_trimer/1'>trimers</scene> and binds <scene name='User:Carolyn_Carr/Sandbox_1/Nisod_monomer/1'>one nickel ion per monomer</scene>. The residues coordinated to the nickel active site are located within the first six N-terminal amino acids, termed the <scene name='User:Carolyn_Carr/Sandbox_1/Nickel-hook/1'>nickel-hook</scene>. The nickel in the active site cycles between Ni(II) and Ni(III). In <scene name='User:Carolyn_Carr/Sandbox_1/Nickel-his_off2/1'>the Ni(II) state</scene> the nickel has a square planar geometry and is coordinated by the side chains of Cys2 and Cys5 as well as the N-terminal amine and the backbone amide group of Cys2. When <scene name='User:Carolyn_Carr/Sandbox_1/Nickel-his_on/1'>oxidized to Ni(III)</scene>, the imidazole group of His1 binds in the axial position forming a square pyramidal geometry. |
| + | {{Clear}} | ||
| + | [[Image:NiSOD Hexamer.png|300 px|thumb|Fig. 2: Hexameric NiSOD. Figure created with PyMol. PDB ID [[1t6u]].]] | ||
| + | {{Clear}} | ||
| + | ===Research Interests=== | ||
| + | ---- | ||
| - | [ | + | The [http://people.chem.umass.edu/mmaroney/research.html | Maroney Lab] is currently investigating the details of the NiSOD catalytic mechanism, including how the enzyme maintains its nickel active sites at 50% Ni(II)/Ni(III) equilibrium despite strong oxidation. In addition, efforts are currently underway to characterize a previously observed intermediate. |
| + | </StructureSection> | ||
| + | ===References=== | ||
| + | ---- | ||
| - | === | + | <references/> |
| + | |||
| + | ===See Also=== | ||
| + | ---- | ||
| - | + | [[1t6i]] <br /> | |
| + | [[1t6q]] <br /> | ||
| + | [[1t6u]] <br /> | ||
| + | [[3g4x]] <br /> | ||
| + | [[3g4z]] <br /> | ||
| + | [[3g50]] | ||
Current revision
| |||||||||||
References
- ↑ Fridovich I. Superoxide dismutases. Annu Rev Biochem. 1975;44:147-59. PMID:1094908 doi:http://dx.doi.org/10.1146/annurev.bi.44.070175.001051
- ↑ Lee JW, Roe JH, Kang SO. Nickel-containing superoxide dismutase. Methods Enzymol. 2002;349:90-101. PMID:11912934
- ↑ Uedsemma, M., Tamm, T. Density-functional theory calculations of aqueous redox potentials of fourth-period transition metals. J. Phys. Chem. A. 2003 Aug 4;107:9997-10003. Epub 2003 Oct 23. 10.1021/jp0362741
- ↑ Bordo D, Matak D, Djinovic-Carugo K, Rosano C, Pesce A, Bolognesi M, Stroppolo ME, Falconi M, Battistoni A, Desideri A. Evolutionary constraints for dimer formation in prokaryotic Cu,Zn superoxide dismutase. J Mol Biol. 1999 Jan 8;285(1):283-96. PMID:9878406 doi:10.1006/jmbi.1998.2267
- ↑ Borgstahl GE, Parge HE, Hickey MJ, Beyer WF Jr, Hallewell RA, Tainer JA. The structure of human mitochondrial manganese superoxide dismutase reveals a novel tetrameric interface of two 4-helix bundles. Cell. 1992 Oct 2;71(1):107-18. PMID:1394426
- ↑ Lah MS, Dixon MM, Pattridge KA, Stallings WC, Fee JA, Ludwig ML. Structure-function in Escherichia coli iron superoxide dismutase: comparisons with the manganese enzyme from Thermus thermophilus. Biochemistry. 1995 Feb 7;34(5):1646-60. PMID:7849024