Molecular Playground/Ubiquitin salt bridge discussion
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
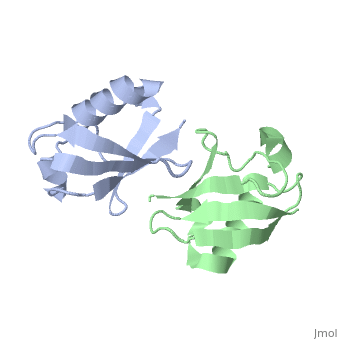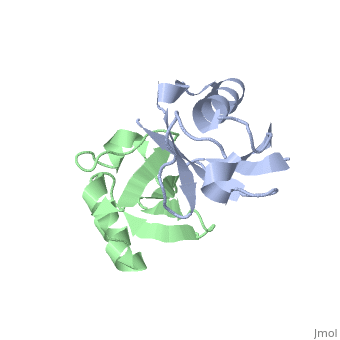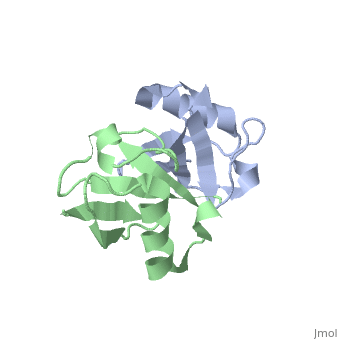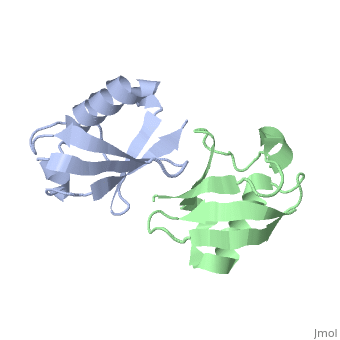
| + | <StructureSection load='2pea' size='350' side='right' scene='' caption='NMR Based Structure of the Closed Conformation of LYS48-Linked Di-Ubiquitin Using Experimental Global Rotational Diffusion Tensor from NMR Relaxation Measurements ([[2pea]])'> | ||
== How native structure is preserved in gas phase == | == How native structure is preserved in gas phase == | ||
| - | {{STRUCTURE_2pea| PDB=2pea | SCENE= }} | ||
| - | |||
This page is going to talk about a gas phase or mass spectrum idea about this. From the perspective of solution phase, we can know the salt bridges stay between E51-R54, R54-D58, E18-K29, D21-K29 and K27-D52. And there is one opinion that most protein when electrosprayed into gas phase from its native solution, the structure features will retain mostly. And electron based dissociation method, like ECD or ETD, can break the protein back bonds instead of disrupting its structure. | This page is going to talk about a gas phase or mass spectrum idea about this. From the perspective of solution phase, we can know the salt bridges stay between E51-R54, R54-D58, E18-K29, D21-K29 and K27-D52. And there is one opinion that most protein when electrosprayed into gas phase from its native solution, the structure features will retain mostly. And electron based dissociation method, like ECD or ETD, can break the protein back bonds instead of disrupting its structure. | ||
Current revision
| |||||||||||
To see the ubiquitin PDB of 2jzz please click .