We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
1x9d
From Proteopedia
(Difference between revisions)
| (16 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | [[Image:1x9d.gif|left|200px]]<br /><applet load="1x9d" size="350" color="white" frame="true" align="right" spinBox="true" | ||
| - | caption="1x9d, resolution 1.410Å" /> | ||
| - | '''Crystal Structure Of Human Class I alpha-1,2-Mannosidase In Complex With Thio-Disaccharide Substrate Analogue'''<br /> | ||
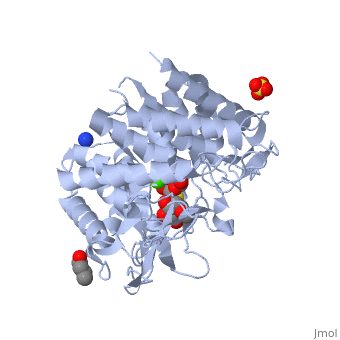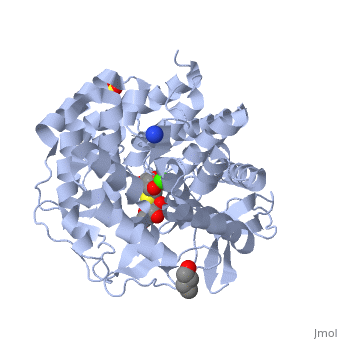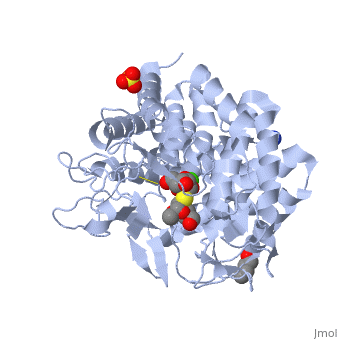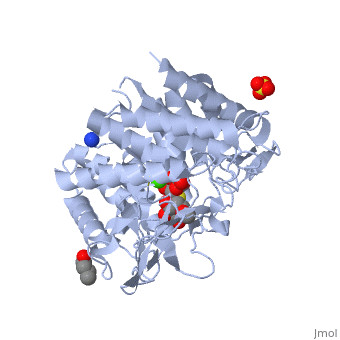
| - | == | + | ==Crystal Structure Of Human Class I alpha-1,2-Mannosidase In Complex With Thio-Disaccharide Substrate Analogue== |
| + | <StructureSection load='1x9d' size='340' side='right'caption='[[1x9d]], [[Resolution|resolution]] 1.41Å' scene=''> | ||
| + | == Structural highlights == | ||
| + | <table><tr><td colspan='2'>[[1x9d]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1X9D OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1X9D FirstGlance]. <br> | ||
| + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 1.41Å</td></tr> | ||
| + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=BU1:1,4-BUTANEDIOL'>BU1</scene>, <scene name='pdbligand=CA:CALCIUM+ION'>CA</scene>, <scene name='pdbligand=MAN:ALPHA-D-MANNOSE'>MAN</scene>, <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene>, <scene name='pdbligand=Z5L:methyl+2-thio-alpha-D-mannopyranoside'>Z5L</scene></td></tr> | ||
| + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1x9d FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1x9d OCA], [https://pdbe.org/1x9d PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1x9d RCSB], [https://www.ebi.ac.uk/pdbsum/1x9d PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1x9d ProSAT]</span></td></tr> | ||
| + | </table> | ||
| + | == Disease == | ||
| + | [https://www.uniprot.org/uniprot/MA1B1_HUMAN MA1B1_HUMAN] Defects in MAN1B1 are the cause of mental retardation autosomal recessive type 15 (MRT15) [MIM:[https://omim.org/entry/614202 614202]. Mental retardation is characterized by significantly below average general intellectual functioning associated with impairments in adaptative behavior and manifested during the developmental period.<ref>PMID:21763484</ref> | ||
| + | == Function == | ||
| + | [https://www.uniprot.org/uniprot/MA1B1_HUMAN MA1B1_HUMAN] Involved in glycoprotein quality control targeting of misfolded glycoproteins for degradation. It primarily trims a single alpha-1,2-linked mannose residue from Man(9)GlcNAc(2) to produce Man(8)GlcNAc(2), but at high enzyme concentrations, as found in the ER quality control compartment (ERQC), it further trims the carbohydrates to Man(5-6)GlcNAc(2).<ref>PMID:12090241</ref> <ref>PMID:18003979</ref> | ||
| + | == Evolutionary Conservation == | ||
| + | [[Image:Consurf_key_small.gif|200px|right]] | ||
| + | Check<jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/x9/1x9d_consurf.spt"</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview03.spt</scriptWhenUnchecked> | ||
| + | <text>to colour the structure by Evolutionary Conservation</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1x9d ConSurf]. | ||
| + | <div style="clear:both"></div> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
Quality control in the endoplasmic reticulum (ER) determines the fate of newly synthesized glycoproteins toward either correct folding or disposal by ER-associated degradation. Initiation of the disposal process involves selective trimming of N-glycans attached to misfolded glycoproteins by ER alpha-mannosidase I and subsequent recognition by the ER degradation-enhancing alpha-mannosidase-like protein family of lectins, both members of glycosylhydrolase family 47. The unusual inverting hydrolytic mechanism catalyzed by members of this family is investigated here by a combination of kinetic and binding analyses of wild type and mutant forms of human ER alpha-mannosidase I as well as by structural analysis of a co-complex with an uncleaved thiodisaccharide substrate analog. These data reveal the roles of potential catalytic acid and base residues and the identification of a novel (3)S(1) sugar conformation for the bound substrate analog. The co-crystal structure described here, in combination with the (1)C(4) conformation of a previously identified co-complex with the glycone mimic, 1-deoxymannojirimycin, indicates that glycoside bond cleavage proceeds through a least motion conformational twist of a properly predisposed substrate in the -1 subsite. A novel (3)H(4) conformation is proposed as the exploded transition state. | Quality control in the endoplasmic reticulum (ER) determines the fate of newly synthesized glycoproteins toward either correct folding or disposal by ER-associated degradation. Initiation of the disposal process involves selective trimming of N-glycans attached to misfolded glycoproteins by ER alpha-mannosidase I and subsequent recognition by the ER degradation-enhancing alpha-mannosidase-like protein family of lectins, both members of glycosylhydrolase family 47. The unusual inverting hydrolytic mechanism catalyzed by members of this family is investigated here by a combination of kinetic and binding analyses of wild type and mutant forms of human ER alpha-mannosidase I as well as by structural analysis of a co-complex with an uncleaved thiodisaccharide substrate analog. These data reveal the roles of potential catalytic acid and base residues and the identification of a novel (3)S(1) sugar conformation for the bound substrate analog. The co-crystal structure described here, in combination with the (1)C(4) conformation of a previously identified co-complex with the glycone mimic, 1-deoxymannojirimycin, indicates that glycoside bond cleavage proceeds through a least motion conformational twist of a properly predisposed substrate in the -1 subsite. A novel (3)H(4) conformation is proposed as the exploded transition state. | ||
| - | + | Mechanism of class 1 (glycosylhydrolase family 47) {alpha}-mannosidases involved in N-glycan processing and endoplasmic reticulum quality control.,Karaveg K, Siriwardena A, Tempel W, Liu ZJ, Glushka J, Wang BC, Moremen KW J Biol Chem. 2005 Apr 22;280(16):16197-207. Epub 2005 Feb 15. PMID:15713668<ref>PMID:15713668</ref> | |
| - | + | ||
| - | + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |
| - | + | </div> | |
| - | + | <div class="pdbe-citations 1x9d" style="background-color:#fffaf0;"></div> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ==See Also== | |
| + | *[[Mannosidase 3D structures|Mannosidase 3D structures]] | ||
| + | == References == | ||
| + | <references/> | ||
| + | __TOC__ | ||
| + | </StructureSection> | ||
| + | [[Category: Homo sapiens]] | ||
| + | [[Category: Large Structures]] | ||
| + | [[Category: Karaveg K]] | ||
| + | [[Category: Liu ZJ]] | ||
| + | [[Category: Moremen KW]] | ||
| + | [[Category: Siriwardena A]] | ||
| + | [[Category: Tempel W]] | ||
| + | [[Category: Wang BC]] | ||
Current revision
Crystal Structure Of Human Class I alpha-1,2-Mannosidase In Complex With Thio-Disaccharide Substrate Analogue
| |||||||||||
Categories: Homo sapiens | Large Structures | Karaveg K | Liu ZJ | Moremen KW | Siriwardena A | Tempel W | Wang BC