This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Jms/sandbox8
From Proteopedia
(Difference between revisions)
(New page: == Your Heading Here (maybe something like 'Structure') == <StructureSection load='1dq8' size='350' side='right' caption='Structure of HMG-CoA reductase (PDB entry 1dq8)' scene=''> Any...) |
|||
| (5 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | == | + | == Prediction about Hsp12 with Gfp == |
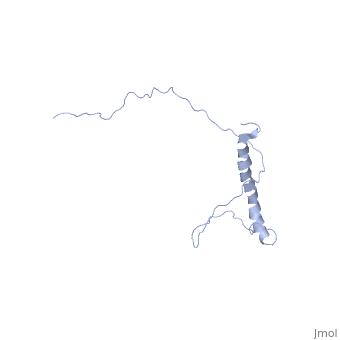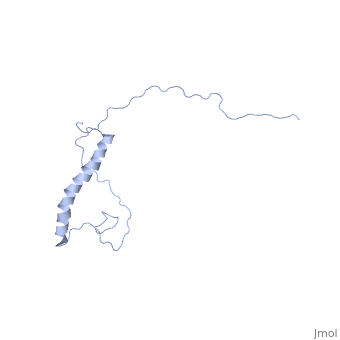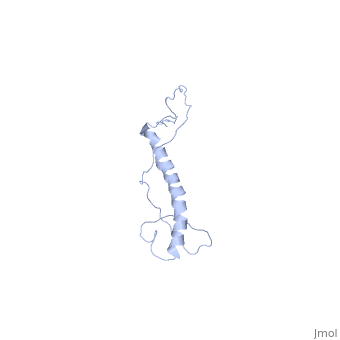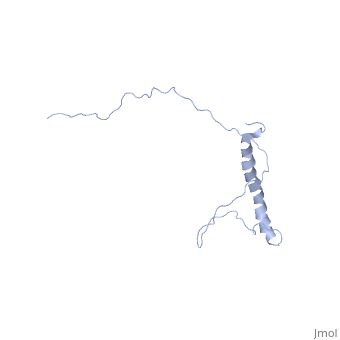
| - | <StructureSection load=' | + | <StructureSection load='2ljl' size='350' side='right' caption='Structure of Hsp12 (PDB entry [[1ljl]])' scene=''> |
| - | + | The GFP likely does not interfere with the native function of Hsp12 because it is far from the localization tags while in the dynamic state in the cytoplasm. And in the membrane it is far from the helixes that actually stabalize the membrane. | |
| - | + | In independent studies Hsp12 was shown to form four helixes upon <scene name='Jms/sandbox8/Hsp12_membrane/1'>localization into the membrane of SDS micelles</scene> | |
| + | which was repeated <scene name='Jms/sandbox8/Hsp12_membrane_2/1'>with the same results</scene> | ||
| + | however, in a dodecaphosphatidylcholine (DPC) micelle, it <scene name='Jms/sandbox8/Hsp12_unfolded/1'>only forms one helix</scene> | ||
</StructureSection> | </StructureSection> | ||
Current revision
Prediction about Hsp12 with Gfp
| |||||||||||