2dpd
From Proteopedia
(Difference between revisions)
| (11 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | [[Image:2dpd.jpg|left|200px]]<br /><applet load="2dpd" size="350" color="white" frame="true" align="right" spinBox="true" | ||
| - | caption="2dpd, resolution 3.17Å" /> | ||
| - | '''Crystal structure of the Replication Termination Protein in complex with a pseudosymmetric B-site'''<br /> | ||
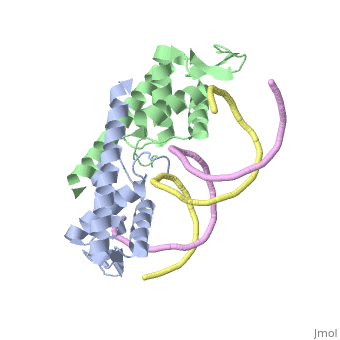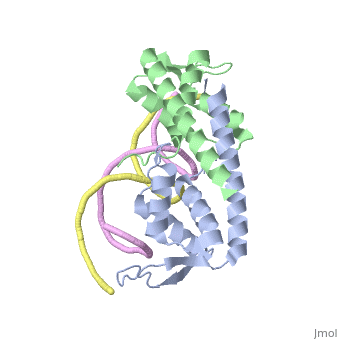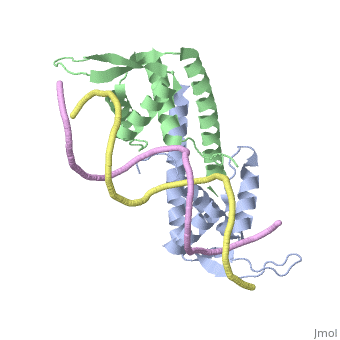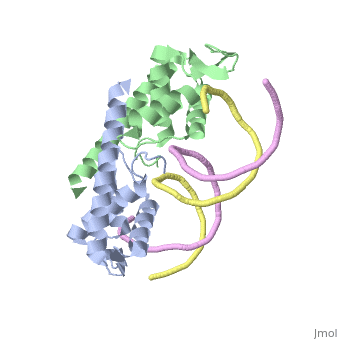
| - | == | + | ==Crystal structure of the Replication Termination Protein in complex with a pseudosymmetric B-site== |
| - | + | <StructureSection load='2dpd' size='340' side='right'caption='[[2dpd]], [[Resolution|resolution]] 3.17Å' scene=''> | |
| - | [ | + | == Structural highlights == |
| - | [[ | + | <table><tr><td colspan='2'>[[2dpd]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/Bacillus_subtilis Bacillus subtilis]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2DPD OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=2DPD FirstGlance]. <br> |
| - | [[ | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 3.17Å</td></tr> |
| - | [ | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2dpd FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2dpd OCA], [https://pdbe.org/2dpd PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2dpd RCSB], [https://www.ebi.ac.uk/pdbsum/2dpd PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2dpd ProSAT]</span></td></tr> |
| - | + | </table> | |
| - | [ | + | == Function == |
| - | + | [https://www.uniprot.org/uniprot/RTP_BACSU RTP_BACSU] Plays a role in DNA replication and termination (fork arrest mechanism). Two dimers of rtp bind to the two inverted repeat regions (IRI and IRII) present in the termination site. The binding of each dimer is centered on an 8 bp direct repeat. | |
| - | [ | + | |
| - | + | ==See Also== | |
| + | *[[Replication Termination Protein|Replication Termination Protein]] | ||
| + | __TOC__ | ||
| + | </StructureSection> | ||
| + | [[Category: Bacillus subtilis]] | ||
| + | [[Category: Large Structures]] | ||
| + | [[Category: Vivian JP]] | ||
| + | [[Category: Wilce J]] | ||
| + | [[Category: Wilce MCJ]] | ||
Current revision
Crystal structure of the Replication Termination Protein in complex with a pseudosymmetric B-site
| |||||||||||