Sandbox Reserved 695
From Proteopedia
(Difference between revisions)
| (3 intermediate revisions not shown.) | |||
| Line 11: | Line 11: | ||
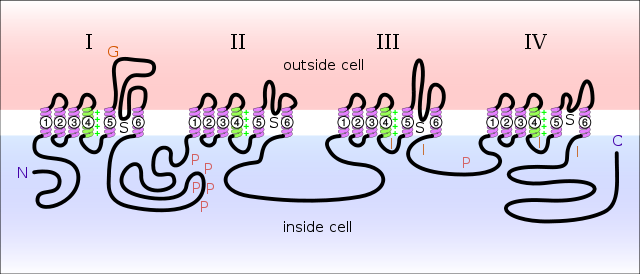
Structural determination of the VGSC has revealed it to be composed of four homologous domains, each of which has six transmembrane regions, while the pore is composed of the S5 and S6 segments <ref>Sato C, Ueno Y, Asai K, Takahashi K, Sato M, Engel A, Fujiyoshi Y: The voltage-sensitive sodium channel is a bell-shaped molecule with several cavities. Nature 2001, 409:1047-1051.</ref>. [[Image:VGSC_SCHEMATIC.png]](Image from Wikimedia<ref>http://en.wikipedia.org/wiki/File:Sodium-channel.svg</ref> For a better illustration of the structure, see [http://genomebiology.com/content/figures/gb-2003-4-3-207-1.jpg Schematic of Transmembrane regions of VGSC] <ref>F.H. Yu, W.A. Catterall Overview of the voltage-gated sodium channel superfamily Genome Biol., 4 (2003), pp. 207–214</ref>.) | Structural determination of the VGSC has revealed it to be composed of four homologous domains, each of which has six transmembrane regions, while the pore is composed of the S5 and S6 segments <ref>Sato C, Ueno Y, Asai K, Takahashi K, Sato M, Engel A, Fujiyoshi Y: The voltage-sensitive sodium channel is a bell-shaped molecule with several cavities. Nature 2001, 409:1047-1051.</ref>. [[Image:VGSC_SCHEMATIC.png]](Image from Wikimedia<ref>http://en.wikipedia.org/wiki/File:Sodium-channel.svg</ref> For a better illustration of the structure, see [http://genomebiology.com/content/figures/gb-2003-4-3-207-1.jpg Schematic of Transmembrane regions of VGSC] <ref>F.H. Yu, W.A. Catterall Overview of the voltage-gated sodium channel superfamily Genome Biol., 4 (2003), pp. 207–214</ref>.) | ||
| - | For better illustration of the structures at work, the structure shown will only contain one subunit. The <scene name='Sandbox_Reserved_695/Vgsc_alphabeta/2'>secondary structure</scene> includes alpha helices | + | For better illustration of the structures at work, the structure shown will only contain one subunit. The <scene name='Sandbox_Reserved_695/Vgsc_alphabeta/2'>secondary structure</scene> includes <font color='blue'>alpha helices</font> and <font color='red'>beta strands</font>. The <scene name='Sandbox_Reserved_695/Vgsc_hydrophobicpolar/2'>hydrophobicity</scene> of the structure reveals that the {{Template:ColorKey_Hydrophobic}} and {{Template:ColorKey_Polar}} residues are not exactly easy to illustrate in their membrane bound and subunit bound region., but the somewhat diffuse polar regions of the molecule are instrumental in providing the channel with enough hydrophillic residues to allow the passage of ions without allowing significant passage of solvent. In coordination with the hydrophobicity illustration, the <scene name='Sandbox_Reserved_695/Vgsc_evolutionaryconservation/2'>evolutionary conservation</scene> plot shows that both some of the hydrophobic and hydrophillic residues are highly conserved {{Template:ColorKey_ConSurf_NoYellow}}. |
==Active Site and Interactions== | ==Active Site and Interactions== | ||
| Line 25: | Line 25: | ||
==References== | ==References== | ||
<references /> | <references /> | ||
| - | |||
| - | Original= <scene name='Sandbox_Reserved_695/Vgsc_orginal/2'>TextToBeDisplayed</scene> | ||
| - | |||
| - | |||
| - | <scene name='Sandbox_Reserved_695/Vgsc_helix_sheet/1'>Alpha Helices and Beta Sheets</scene> | ||
| - | |||
| - | |||
| - | <scene name='Sandbox_Reserved_695/Vgsc_ligand_residues/1'>Ligand Binding</scene> | ||
| - | <scene name='Sandbox_Reserved_695/Vgsc_ligand_spacefill/1'>Ligand binding Space-fill</scene> | ||
Current revision
| This Sandbox is Reserved from 30/01/2013, through 30/12/2013 for use in the course "Biochemistry II" taught by Hannah Tims at the Messiah College. This reservation includes Sandbox Reserved 686 through Sandbox Reserved 700. |
To get started:
More help: Help:Editing |
Epileptic drugs at Voltage-Gated Sodium Ion Channel
Introduction
| |||||||||||